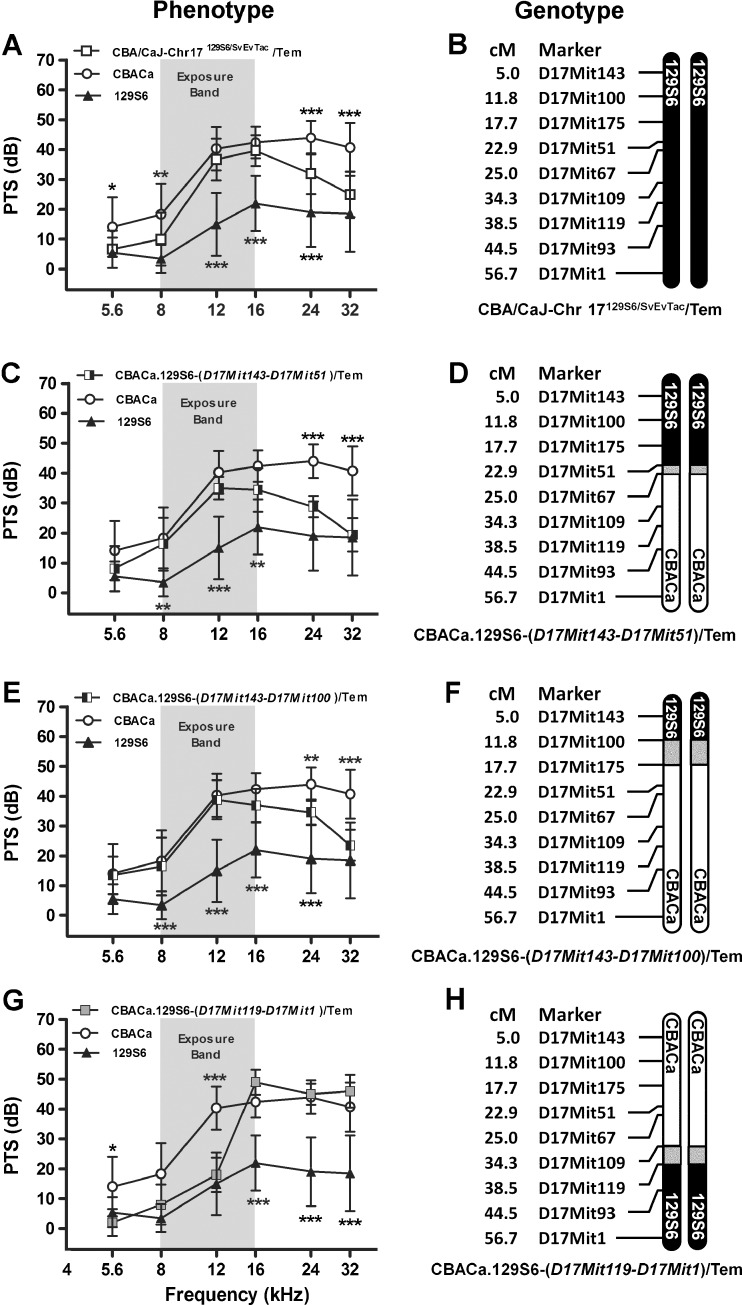
FIG. 9.
Chromosome 17 congenic strains. PTS (A, C, E, G) for three Chr 17 congenic strains are compared to the parental CBACa (n = 29) and 129S6 (n = 10) strains, displayed with the recombinant crossover event defining each strain (B, D, F, H). Other display conventions are as described for Figure 8. A 129S6 Chr 17 appears to contribute to NR at 5.6, 8, 24, and 32 kHz in the CBA/CaJ-Chr 17129S6/SvEvTac/Tem strain (n = 15). PTS data are the same as those shown in Figure 7C, duplicated here to allow comparison among all three congenic strains generated from the CBA/CaJ-Chr 17129S6/SvEvTac/Tem consomic strain. B This consomic strain contains a nonrecombinant 129S6 Chr 17. C The CBACa.129S6-(D17Mit143-D17Mit51)/Tem (n = 8) strain demonstrates NR at 24 and 32 kHz. The overall strain effect in the ANOVA is F2,264 = 100, p < 0.0001. D This congenic strain contains a crossover between D17Mit51 and D17Mit67. E The CBACa.129S6-(D17Mit143-D17Mit100)/Tem strain (n = 13) demonstrates NR at 24 and 32 kHz. The overall strain effect in the ANOVA is F2,294 = 133, p < 0.0001. F This congenic strain contains a crossover between D17Mit100 and D17Mit175. G The CBACa.129S6-(D17Mit119-D17Mit1)/Tem strain (n = 5) demonstrates significant NR at 5.6 and 12 kHz. H This congenic strain is defined by a crossover between D17Mit109 and D17Mit119. The overall strain effect in the ANOVA is F2,246 = 82, p < 0.0001.