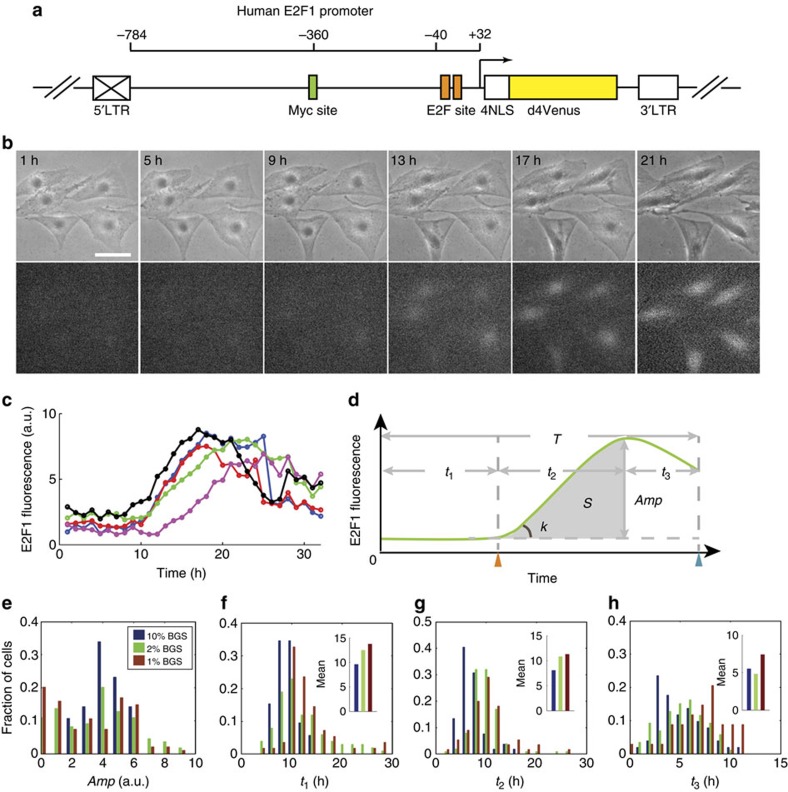
Figure 2. Quantification of E2F dynamics in single cells.
(a) hE2F1p::4NLS-d4Venus reporter construct. Box with cross indicates that 5′ long terminal repeat (LTR) is inactivated after viral integration. (b) Time-lapse microscopy images of REF52 rat fibroblasts expressing hE2F1p::4NLS-d4Venus reporter released from serum starvation by adding 10% bovine growth serum (BGS). Upper panel, phase channel; low panel, Venus channel. Scale bar, 50 μm. (c) E2F1 dynamics trajectories of the five individual cells shown in b. Time-series raw data (sampled per hour) were smoothened in a 3-h time window. (d) Characterization of E2F1 dynamics trajectory in divided (green) cells with defined metrics: t1, initial delay; t2, activation time; t3, post-activation time; Amp, amplitude; S (shadow area), total E2F1 work during activation time; k, slope; T, the entire cell cycle length. Orange triangle indicates E2F1 signal upturn time point and the blue one indicates cell division time point. (e) Histogram shows the distribution of Amp at different serum levels. (f–h) Histograms show distributions of t1, t2 and t3 (in divided cells) at different serum levels. The mean values were compared among different serum concentrations and plotted as insets.