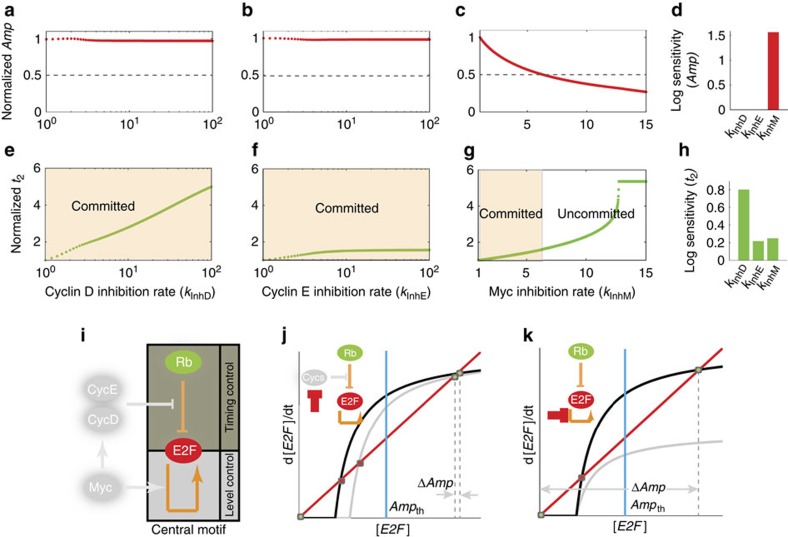
Figure 4. Modelling predicts distinct modes of regulation on E2F dynamics by G1 cyclins and Myc.
(a–c) Normalized Amp (to the Amp for base parameter values) within 120 h under the inhibition of CDK4/6, CDK2 or Myc as function of inhibition rate. Horizontal dashed lines mark that the expected threshold value for normalized Ampth is 0.5, given that the measured Ampth is around half of the average Amp of cells (2.5 versus 5, Fig. 3e). (d) Log sensitivity analysis of Amp to inhibition rate. (e–g) Normalized t2 to its initial level as a function of inhibition rate under each inhibition case. Yellow shading highlights the region where cells are expected to commit into cell cycle. (h) Log sensitivity analysis of t2 to inhibition rate. For kInhM, log sensitivity was calculated within the committed region. (i) The central motif of Myc/Rb/E2F network includes E2F auto-regulation (module for level control) and its titration by Rb (module for timing control). (j) Steady-state analysis of the central motif under the inhibition of Rb phosphorylation through reduction of cyclin complexes activity. The decrease in Rb phosphorylation rate leads to only a slight reduction (ΔAmp) in steady-state E2F level. Black and grey lines are E2F synthesis curves under low (black) and high (grey) cellular unphosphorylated Rb levels, respectively. Red line indicates the degradation curve. Blue line indicates the threshold Ampth. Green dots, stable steady states; red dots, unstable steady states. (k) Steady-state analysis of the central motif. Black and grey lines are E2F synthesis curves given high (black) and low (grey) synthesis rate, respectively.