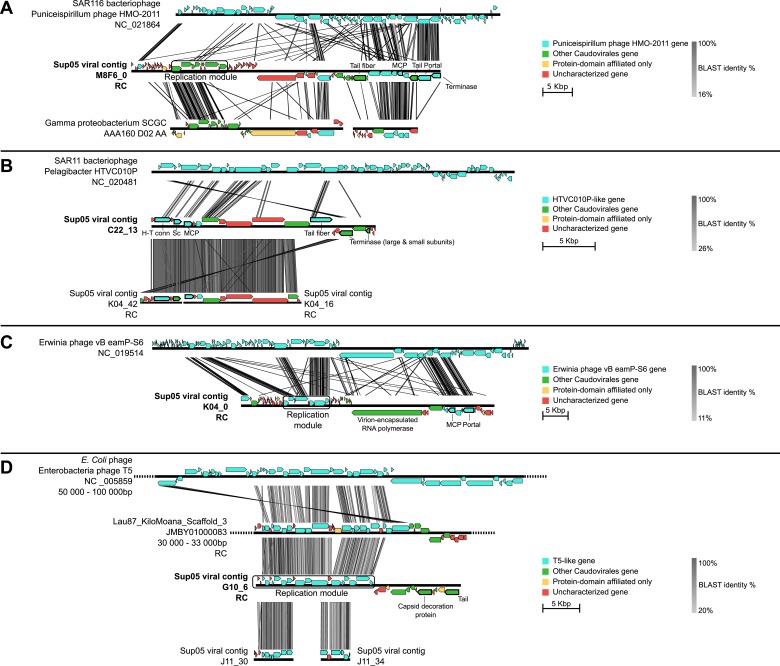
Figure 2. Genetic map and synteny plots for the four references SUP05 Caudovirales contigs M8F6_0 (A), C22_13 (B), K04_0 (C) and G10_6 (D) (highlighted in bold).
Viral hallmark genes are underlined and identified on plots (MCP: major capsid protein, Sc: scaffolding protein, H-T conn.: head-tail connector). Sequence similarities were deduced from a tBLASTx comparison. For clarity sake, several sequences including SUP05 viral contig M8F6_0, K04_0, and G10_6 are reverse-complemented (noted RC).
DOI: http://dx.doi.org/10.7554/eLife.03125.008