Abstract
BACKGROUND
Epstein-Barr virus (EBV) persists in infected B lymphocytes in blood donors. Lymphocytes are viable during platelet (PLT) storage. The effects of storage and leukoreduction on lymphocytes and EBV genomes are evaluated.
STUDY DESIGN AND METHODS
Forty nonleukoreduced PLT concentrates were stored at 20 to 24°C for up to 7 days. EBV genomes in B cells were quantified on Days 1 and 5. Viable white blood cells (WBCs) and T and B cells were quantified in 10 of 40 units on Days 1, 3, 5, and 7 of storage. For the leukoreduction study, four pools of PLTs were leukoreduced within 24 hours of collection. B cells from before leukoreduction and all peripheral blood mononuclear cells from after leukoreduction were assayed for EBV.
RESULTS
Viable WBCs and T cells were stable whereas viable B cells were reduced to 71% of the Day 1 level by Day 5. A total of 31 of 37 (83.8%) units were EBV positive. Although EBV genomes remained stable in most units, 12 of 37 units demonstrated a median of 5.1 (range 2- to 134)-fold increase in EBV genomes per 105 B cells on Day 5. For the leukoreduction study, EBV genomes were detected in four of four pools before leukoreduction with a median of 3.8 (range, 0.2–93.6) EBV genomes per 105 B cells. EBV genomes were not detected in any of the postleukoreduction specimens.
CONCLUSIONS
Seventy percent of B lymphocytes are viable on Day 5 of PLT storage. Although the mean number of EBV genomes remained stable, a subset of units had increased EBV genomes during storage. Leukoreduction removed polymerase chain reaction–detectable EBV genomes from PLT pools.
Epstein-Barr virus (EBV) infects and establishes lifelong latency in peripheral blood B lymphocytes in a majority of the adult population, including blood donors.1,2 The frequency of EBV-infected circulating B lymphocytes is estimated to be 1 in 105 to 1 in 106 in normal latency in healthy individuals.3,4 In an immunocompromised host, EBV is implicated in the etiology of Burkitt’s lymphoma and lymphoproliferative disorders. We have previously described the effects of leukoreduction and storage on EBV genomes in RBC components.5,6 In this study we investigated the presence and stability of EBV genomes in platelet (PLT) concentrates, which were stored in standard blood bank conditions and the effect of leukoreduction on EBV removal.
MATERIALS AND METHODS
Whole-blood PLT processing
For the storage study, 40 units of nonleukoreduced whole-blood PLT concentrates randomly selected from the regional FDA-licensed blood center (Central Blood Bank, Pittsburgh, PA) were stored under standard blood bank conditions at 20 to 24°C for up to 7 days. The volume of the PLT concentrates ranged from 66 to 74 mL with a mean of 69.2 mL. A sterile docking device was used to aliquot 1-mL specimens from 10 of the 40 PLTs concentrates on Days 1, 3, 5, and 7 of storage to assess cell content and viability by flow cytometry. A 20- to 24-mL specimen was removed on Day 1 of storage and the remaining volume was stored for 5 days to quantify EBV genomes.
For the leukoreduction study, four pools (five PLT concentrates in each pool) of PLTs were processed within 24 hours of collection. The volume of the pools before leukoreduction ranged from 359 to 373 mL with a mean of 367.2 mL. Fifty milliliters was taken from each pool before filtration for CD19+ B-cell purification to detect EBV DNA. The remainder of the pools underwent leukoreduction with a leukoreduction filter (PXL-8, Pall, East Hills, NY) according to the manufacturer’s instructions. The entire volume from each leukoreduced pool was used for peripheral blood mononuclear cell (MNC) isolation and EBV DNA detection.
Assessment of lymphocyte viability
Total WBCs (CD45+), T lymphocytes (CD3+), and B (CD19+ and CD20+) lymphocytes were quantified by a single-platform flow cytometric assay in 10 PLT concentrates on Days 1, 3, 5, and 7 of storage using previously described methods.6 Each specimen (0.1 mL of PLTs) was stained in duplicate with anti-CD45–fluorescein isothiocyanate (FITC) plus CD3-phycoerythrin (PE) or CD19-PE plus CD20 PE (Beckman Coulter, Fullerton, CA) for WBCs, T cells, and B cells, respectively. A negative control cocktail consisting of anti-CD45–FITC/immunoglobulin G1–PE was performed in a third tube. Viability was assessed by 7-aminoactinomycin D exclusion. Data were acquired on a four-color cytometer (Epics XL, Beckman Coulter) calibrated daily with beads (Flow Check and Flow Set, Beckman Coulter). Blood reference cells (CD-Chex Plus, Streck Laboratories, Omaha, NE) were run daily as a positive control for CD3 and CD19. Raw data were organized in a computer spreadsheet (Excel, Microsoft Corp., Redmond, WA) and analyzed using computer software (Systat v11, Systat Software, Inc., Richmond, CA).
MNC fractionation and CD19+ cell selection
MNCs were isolated from all specimens by Ficoll density gradient centrifugation. PLT concentrate specimens were mixed with equal volume of phosphate-buffered saline (1× PBS, pH 7.4, BioWhittaker, Rockland, ME) and then layered onto Ficoll-Hypaque (Histo-paque 1077, Sigma Diagnostics, St Louis, MO). After centrifugation, the interface layer was carefully aspirated. MNCs were washed twice with PBS. To acquire B lymphocytes from all specimens in the PLT storage study and preleukoreduced specimens in the leukoreduction study, MNCs were incubated with 4:1 ratio of CD19+ micro-beads (Miltenyi Biotec, Auburn, CA) for 30 minutes at 6°C. CD19+ B cells were collected with an LS separation column (Miltenyi Biotec, Auburn, CA) and stored as a pellet at −70°C until use. For leukoreduction pools, MNCs from the entire pool were harvested and stored as a cell pellet at −70°C until use.
Quantification of EBV genomes by real-time polymerase chain reaction
DNA was extracted from the purified B-cell pellet (all specimens in the PLT storage study and preleukoreduced specimens in the leukoreduction study) and the MNC cell pellet (postleukoreduced specimens in leukoreduction study) using a genomic DNA purification kit (Gentra System, Minneapolis, MN). EBV quantification was performed by amplifying a DNA target in the EBV latent membrane protein 2a (LMP2a) gene using a real-time polymerase chain reaction (PCR) method described previously.5 A DNA PCR amplifying cellular GAPDH gene involved in the glycolytic pathway was run in parallel to determine the input cell number for EBV PCR.5 All standards, controls, and samples were run in duplicates for both EBV and GAPDH DNA quantification except for postleukoreduced specimens. DNA extracted from the MNC fraction from each leukoreduced PLT pool was used in a single PCR procedure to enhance detection sensitivity; approximately 4% of the DNA was used in a separate PCR for MNC quantification. Amplification and data acquisition were run on a sequence detection system (ABI 7700, Applied Biosystems, Foster City, CA). The coefficient of variation of intra- and interassay were below 20% as previously reported.5,6 The EBV genomes per 105 B cells were calculated based on results from the PCR procedures in which DNA equivalent to 106 cells was used in the amplification.
RESULTS
Viability of WBCs, T cells, and B cells in PLT concentrates
The median (95% confidence interval [CI]) viable WBC, T-cell, and B-cell counts in PLT concentrates over a 7-day period are summarized in Table 1. There were no significant changes in the number of viableWBCs and T lymphocytes during PLT storage for up to 7 days at 20 to 24°C. For instance, the median number of T cells was 324 on Day 1 and 320 on Day 7. In contrast, the median number of viable B cells on Day 1 was 62 and that number was reduced to 44 on Day 5, a 30% drop. In only one PLT concentrate was there an incremental increase in B cells from 37 (cells/μL) on Day 1 to 44 on Day 3, 56 on Day 5, and 73 on Day 7, a 197% increase in cell number from Day 1 to Day 7. Over the same period, the T-cell counts also increased from 620 (cells/μL) on Day 1 to 864 on Day 5 and 1077 on Day 7, the latter being notable because it was the highest T-cell count detected in any unit at any time.
TABLE 1.
EBV genomes and viable WBCs, T and B cells during PLT storage*
| Day | EBV genomes (n = 10) | WBCs (n = 10) | T cells (n = 10) | B cells (n = 10) |
|---|---|---|---|---|
| 1 | 1.6 (0.1–15.8) | 591 (380–802) | 324 (187–324) | 62 (30–94) |
| 3 | Not tested | 498 (322–672) | 272 (164–382) | 53 (28–80) |
| 5 | 4.3 (0.3–17.6) | 571 (329–814) | 341 (174–507) | 44 (22–65) |
| 7 | Not tested | 603 (289–917) | 320 (114–526) | 32 (16–48) |
EBV genomes are reported as median (range) of genomes/105 B cells; WBCs, T cells, and B cells are reported as median (95% CI) viable cells/μL.
EBV genomes in CD19+ B lymphocytes during PLT storage
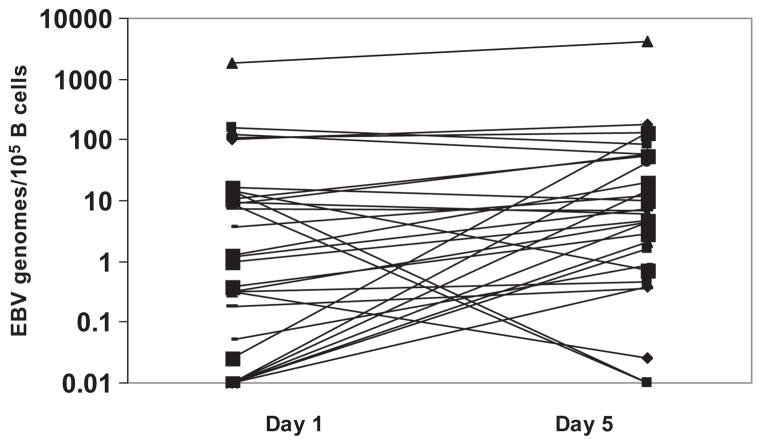
Of 40 PLT concentrates, 3 could not be analyzed due to insufficient DNA obtained either on Day 1 or on Day 5 of storage. EBV DNA was detected in 31 of 37 (83.8%) evaluated specimens. The median (range) values of EBV genomes in the 31 DNA positive specimens were 1.15 (0.1–1835.1) and 6.2 (0.1–4196.9) genomes per 105 B cells on Days 1 and 5 of storage, respectively. One PLT concentrate contained very high level of EBV genomes of approximately 1000-fold of the median viral load. Sixteen of 37 had EBV genomes less than a twofold increase or decrease between Day 1 and Day 5. In 3 of 37 units, EBV genomes decreased by 11-fold (range, 8- to 14-fold). Twelve of 37 specimens produced median (range) EBV genomes increase of 5.1-fold (2- to 134-fold). Figure 1 shows EBV genomes in 31 DNA-positive specimens on Days 1 and 5 of storage. Of the 10 PLT concentrates for which detailed flow cytometric analysis of B and T cells were made (Table 1), 4 had an increase in EBV genomes between Day 1 and Day 5 of storage. The fold increases were 2.7, 5.4, 43, and 4.4 for PLT concentrate (units) 3, 6, 8, and 9 in Table 2. All four showed a decrease in the numbers of both B and T cells (Table 2). The decreases in cell counts were similar to that observed in the other six PLT concentrates that did not have more than twofold changes in EBV genomes from Day 1 to Day 5 of storage (not shown).
Fig. 1.
EBV genomes in CD19+ B lymphocytes during PLT storage. Graph shows changes in EBV genomes on Days 1 and 5 of storage for 31 of 37 EBV-positive PLT concentrates. Sixteen of 37 had EBV genomes less than a twofold increase or decrease. In 3 of 37 units, EBV genomes decreased by 11-fold (range, 8- to 14-fold). Twelve of 37 specimens produced median (range) EBV genomes increase of 5.1-fold (2- to 134-fold).
TABLE 2.
Measurements of B and T cells in PLT units showing an increase in EBV
| B cells (/μL)
|
T cells (μL)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Unit | Day 1 | Day 3 | Day 5 | Day 7 | Unit | Day 1 | Day 3 | Day 5 | Day 7 |
| 3 | 56 | 63 | 40 | 35 | 3 | 444 | 371 | 373 | 293 |
| 6 | 13 | 11 | 8 | 9 | 6 | 85 | 80 | 76 | 111 |
| 8 | 132 | 96 | 75 | 50 | 8 | 437 | 326 | 433 | 313 |
| 9 | 113 | 111 | 97 | 53 | 9 | 530 | 493 | 510 | 463 |
Effects of leukoreduction on EBV genomes in PLT concentrates
The median number of MNCs for preleukoreduced five-pool PLT concentrates was 2.3 × 108, ranging from 1.6 × 108 to 6.2 × 108. EBV DNA was detected in all four pools with median (range) of 3.9 (0.2–93.6) EBV genomes per 105 B cells. This translates to median (range) EBV genomes of 1392 (43–16,842) in the pools, calculated from the total number of viable B cells in the preleukoreduced specimens. The median number of MNCs recovered from the entire pool of the postleukoreduced specimens was 2.1 × 104 (range, 1.3 × 104–4.0 × 104). No EBV DNA was detected in any of the four pools after leukoreduction.
DISCUSSION
We have previously reported the effects of leukoreduction and storage on EBV genomes during RBC storage.5,6 Leukoreduction removed 4-log EBV genomes from the RBC units and rendered most RBC units EBV negative by PCR.5 The RBC storage study indicated that 19% of B lymphocytes were viable at the end of RBC storage and EBV latently infected B cells can survive the normal storage conditions for RBCs.6 In this study, we applied the same methods to evaluate the effect of storage on viable B cells and EBV genomes and efficacy of EBV removal through leukoreduction in nonleukoreduced PLT concentrates.
Most WBCs, T lymphocytes, and B lymphocytes in the PLT products remain viable for up to 7 days of the storage at 20 to 24°C. Viable lymphocytes in blood products are capable of replicating and engrafting when transfused to immunocompetent transfusion recipients resulting in chimerisim.7 Unlike RBC storage at 1 to 6°C under which lymphocytes are unable to proceed through the cell cycle, the storage conditions for PLTs (20–24°C) could potentially permit lymphocytes to replicate during storage. This study was unable to evaluate whether lymphocytes proliferated during PLT storage.
EBV genomes were detected in 84% of PLT concentrates, reflecting the high prevalence of EBV in blood donors and the general population. The median number of EBV genomes per 105 B cells was one and six on Days 1 and 5 of storage, respectively. These results are in agreement with our previous observations.5,6,8 Similar to our previous observation in the RBC study,6 a single whole-blood PLT concentrate contained very high EBV genome level almost 1000-fold higher than is normally associated with EBV latency. The health status of this donor is unknown due to lack of linkage information. There was a median 5-fold (range, 2- to 134-fold) increase in EBV genomes on Day 5 of PLT storage (compared to Day 1) in 12 of 37 EBV-positive PLT concentrates. This could be due to either an increase in the number of EBV-infected cells (cell replication) or an increase in the number of EBV genomes per infected cell (viral replication), or both, since the storage temperature of the PLT potentially permits both events to occur. This study was unable to determine the exact mechanism.
As would be expected and has been demonstrated previously, leukoreduction effectively removed EBV genomes from all four pools of PLTs concentrates, rendering the filtered products negative by PCR for B-cell-associated EBV. The current study did not evaluate whether free viral particles produced by possible lytic reactivation of the latently infected B cells upon in vitro storage, were present. Transmission of EBV through transfusion of nonleukoreduced blood products has been discussed previously by us5 and others.9 Documentation of EBV transmission from an EBV-positive blood donor to an EBV-negative 16-year-old liver transplant recipient (who received an organ from an EBV-negative organ donor) was accomplished by DNA analysis and serology.10 The blood donor in the case had clinical infectious mononucleosis 15 months before the donation. In summary, although the EBV genomes persist through PLT storage, leukoreduction provides an effective means of removing cell-associated EBV genomes. Although this is not a clinical study of EBV transmission, EBV PCR–negative blood products are likely to have a very low probability of viral transmission. These data provided a scientific basis to support the efficacy of leukoreduction in reducing the risk of EBV transmission from PLTs.
ABBREVIATION
- EBV
Epstein-Barr virus
Footnotes
CONFLICT OF INTEREST
The authors claim no conflict of interest.
References
- 1.Klein G. Epstein-Barr virus strategy in normal and neoplastic B cells. Cell. 1994;77:791–3. doi: 10.1016/0092-8674(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 2.Qu L, Rowe DT. Epstein-Barr virus latent gene expression in uncultured peripheral blood lymphocytes. J Virol. 1992;66:3715–24. doi: 10.1128/jvi.66.6.3715-3724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyashita EM, Yang B, Babcock GJ, Thorley-Lawson DA. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell [erratum appears in J Virol 1998 Nov;72(11):9419] J Virol. 1997;71:4882–91. doi: 10.1128/jvi.71.7.4882-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babcock GJ, Decker LL, Volk M, Thorley-Lawson DA. EBV persistence in memory B cells in vivo. Immunity. 1998;9:395–404. doi: 10.1016/s1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- 5.Qu L, Xu S, Rowe D, Triulzi D. Efficacy of Epstein-Barr virus removal by leukoreduction of red blood cells. Transfusion. 2005;45:591–5. doi: 10.1111/j.0041-1132.2005.04303.x. [DOI] [PubMed] [Google Scholar]
- 6.Qu L, Triulzi DJ, Rowe DT, Griffin DL, Donnenberg AD. Stability of lymphocytes and Epstein-Barr virus during red blood cell storage. Vox Sang. 2007;92:125–9. doi: 10.1111/j.1423-0410.2006.00880.x. [DOI] [PubMed] [Google Scholar]
- 7.Lee TH, Paglieroni T, Ohto H, Holland PV, Busch MP. Survival of donor leukocyte subpopulations in immunocompetent transfusion recipients: frequent long-term microchimerism in severe trauma patients. Blood. 1999;93:3127–39. [PubMed] [Google Scholar]
- 8.Rowe DT, Qu L, Reyes J, Jabbour N, Yunis E, Putnam P, Todo S, Green M. Use of quantitative competitive PCR to measure Epstein-Barr virus genome load in the peripheral blood of pediatric transplant patients with lymphoproliferative disorders. J Clin Microbiol. 1997;35:1612–5. doi: 10.1128/jcm.35.6.1612-1615.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillyer CD, Lankford KV, Roback JD, Gillespie TW, Silberstein LE. Transfusion of the HIV-seropositive patient: immunomodulation, viral reactivation, and limiting exposure to EBV (HHV-4), CMV (HHV-5), and HHV-6, 7, and 8. Transfus Med Rev. 1999;13:1–17. doi: 10.1016/s0887-7963(99)80084-4. [DOI] [PubMed] [Google Scholar]
- 10.Alfieri C, Tanner J, Carpentier L, Perpête C, Savoie A, Paradis K, Delage G, Joncas J. Epstein-Barr virus transmission from a blood donor to an organ transplant recipient with recovery of the same virus strain from the recipient’s blood and oropharynx. Blood. 1996;87:812–7. [PubMed] [Google Scholar]