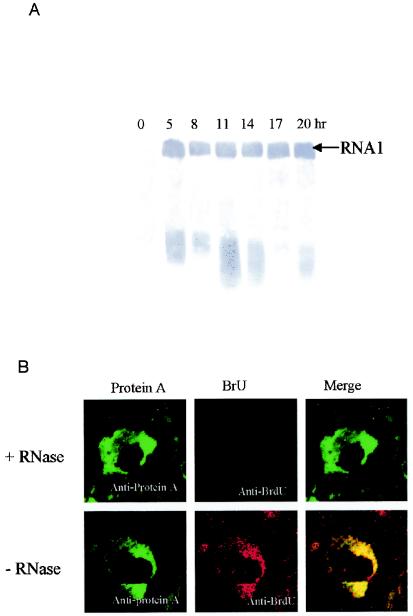
FIG. 4.
(A) Temporal pattern of GGNNV RNA1 replication in infected SB cells. Total RNA of SB cells infected with GGNNV was extracted at 3-h intervals from 5 h p.i. up to 20 h p.i. Total RNA was separated on denatured formaldehyde-agarose gels, transferred to a nylon membrane, and blotted with DIG-labeled probe that detected positive-strand RNA1. (B) Colocalization of GGNNV protein A with nascent viral RNA synthesis in infected SB cells. GGNNV-infected SB cells were BrUTP labeled at 14 h p.i. for 1 h and dual labeled with anti-protein A sera (red) and anti-BrdU MAb (green). Cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and immunostained. As a specificity control, BrUTP-labeled cells were treated with RNase after fixation and permeabilization but before immunostaining (top). The merged images represent digital superimposition of green and red signals, where areas of fluorescence colocalization are yellow-orange. Magnification, ×630.