Table 1.
Testing for an initial cline pattern in 15 microsatellites
| MfSSR | 407 | 425 | 405 | 322 | 324 | 413 | 434 | 340 | 401 | 430 | 362 | 417 | 428 | 203 | 350 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LLT1 (k = 5) | −84.42 | −46.74 | −87.17 | −83.71 | −74.65 | −85.89 | −83.46 | −82.19 | −38.45 | −16.41 | −16.62 | −88.39 | −91.55 | −75.26 | −36.14 |
| θ* | 1.871 | 1.878 | 1.869 | 1.870 | 1.870 | 1.875 | 1.881 | 1.873 | x | x | x | x | x | x | x |
| c (km)* | 20.06 | 20.08 | 19.87 | 21.07 | 20.09 | 21.02 | 19.94 | 19.99 | x | x | x | x | x | x | x |
| b* | 68.33 | 140.19 | 11.20 | 25.73 | 17.25 | 35.72 | 38.97 | 22.92 | 0.10 | 0.03 | 0.07 | 0.09 | 0.12 | 0.02 | 0.06 |
| h1* | 0.13 | 0.62 | 0.00 | 0.08 | 0.02 | 0.16 | 0.17 | 0.00 | 0.67 | 0.99 | 0.96 | 0.45 | 0.46 | 0.12 | 0.85 |
| hp* | 0.59 | 0.94 | 0.95 | 0.64 | 0.98 | 0.69 | 0.78 | 0.70 | 0.01 | 1.00 | 1.00 | 0.05 | 0.00 | 0.10 | 0.04 |
| h2† | 0.64 | 0.98 | 0.95 | 0.67 | 0.98 | 0.74 | 0.82 | 0.70 | 0.67 | 1.00 | 1.00 | 0.47 | 0.46 | 0.21 | 0.85 |
| Slope† | 8.79 | 12.62 | 2.67 | 3.77 | 4.16 | 5.24 | 6.29 | 3.99 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Width (km)† | 6.44 | 3.14 | 39.30 | 17.10 | 25.51 | 12.32 | 11.29 | 19.20 | x | x | x | x | x | x | x |
| LL0 Worse model (k = 1) | −95.54 | −59.02 | −93.36 | −92.43 | −84.69 | −94.48 | −96.29 | −91.61 | −38.46 | −18.13 | −18.13 | −88.95 | −93.19 | −76.76 | −37.14 |
| Dev: −2LL (LL0–LLT1) | 22.24 | 24.55 | 12.40 | 17.44 | 20.07 | 17.17 | 25.65 | 18.84 | 0.02 | 3.46 | 3.04 | 1.11 | 3.29 | 3.01 | 2.00 |
Clines were fitted using scaled Logit function (Equation in Appendix S6).
Denotes inferred parameters: cline centre (c), cline slope-related parameter (b), lower asymptotic frequency (h1), allelic frequency step between the two populations (hp) and cline angle (θ).
Holds for a posteriori computed parameters. These are h2, the higher asymptotic frequency 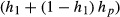 , cline slopes
, cline slopes 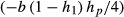 and for non-flat clines, cline widths (4/b, in km). LL is log likelihood. For each locus, slope significance was tested using likelihood ratio tests against a null model assuming homogeneous allele frequency over the study area. Deviance (Dev) is given with significant values (5% level) highlighted in bold × indicates situations in which parameters could not be computed (see text for details).
and for non-flat clines, cline widths (4/b, in km). LL is log likelihood. For each locus, slope significance was tested using likelihood ratio tests against a null model assuming homogeneous allele frequency over the study area. Deviance (Dev) is given with significant values (5% level) highlighted in bold × indicates situations in which parameters could not be computed (see text for details).
