FIG. 8.
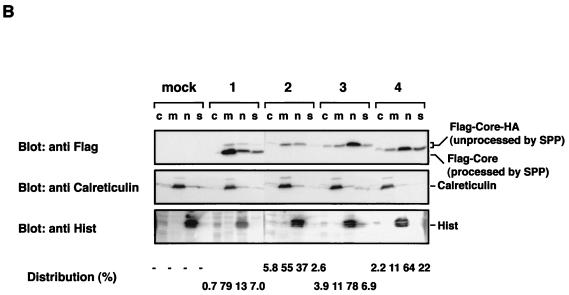
Localization of mutant HCV core proteins. (A) Putative processing mechanisms of wild-type and mutant core proteins is illustrated at the top. EGFP-Core-191 (column 1), EGFP-Core IF176/177AL (column 2), EGFP-Core LVL/3A (column 3), and EGFP-Core Δ128-151 (column 4) were coexpressed with ER-DsRed in HeLa cells, and subcellular localization of core proteins was examined by confocal microscopy. (B) Subcellular fractionation of HeLa cells transfected with plasmids encoding Flag-Core-HA polyproteins. Cells transfected with an empty plasmid (lane M) or plasmid encoding Flag-Core 191-HA (lanes 1), Flag-Core IF176/177AL-HA (lanes 2), Flag-Core LVL/3A-HA (lanes 3), or Flag-Core Δ128-151-HA (lanes 4) were extracted into four fractions, as described in Materials and Methods. Each fraction was concentrated and subjected to immunoblotting with anti-Flag antibody (upper panel). Lanes c, m, n and s, cytosol, membrane-organelle, nuclear, and cytoskeleton fractions, respectively. Calreticulin and histone (His) were used as markers for membrane-organelle and nuclear fractions, respectively. To determine the distributed ratio of processed and unprocessed core proteins in each fraction, the density of core protein in each fraction was measured and is indicated as a percentage at each bottom of lane.