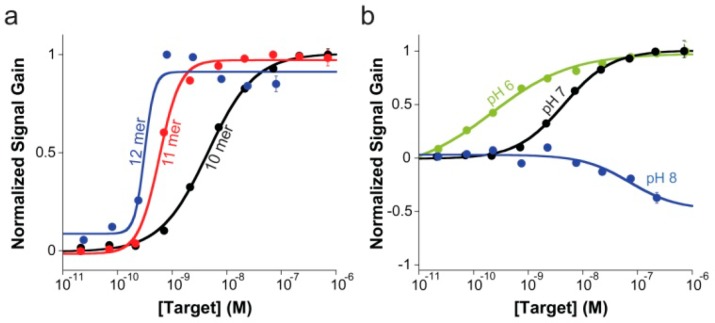
Figure 4.
(a) E-DNA clamp-switch sensor can detect specific complementary targets with high affinity. Here are shown binding curves obtained by using increasing concentration of complementary targets of different lengths (10, 11, and 12 bases). As expected, the affinity observed with longer targets is improved until we reach the ligand-depletion regime in which occupancy is no longer defined by the true affinity of the probe or the concentration of the target in solution but by the total number of ligand (target) molecules in the sample relative to the total number of probes on the sensor surface.24 In this latter case, a bilinear binding curve is observed with a midpoint at a target concentration half of the effective probe concentration ([P]eff/2). These binding curves were obtained by adding an increasing concentration of perfectly matched targets of different length in a 2 mL 10 mM TRIS buffer, 10 mM MgCl2, 100 mM NaCl pH 7.0. (b) Sensing mechanism of the E-DNA clamp-switch sensor is based on the formation of a triplex structure upon target binding. Consistent with this and considering that triplex formation is unfavored at basic pH,4,14−17 the affinity of our clamp-switch sensor becomes poorer as we increase the pH at which we interrogate the sensor. Interestingly, because a basic pH (here pH 8.0, blue curve) greatly inhibits triplex formation, we only observe duplex formation. These binding curves were obtained by adding an increasing concentration of a perfectly matched target (10-mer) in a 2 mL 10 mM TRIS buffer, 10 mM MgCl2, 100 mM NaCl (pH 6, 7 and 8).