Figure 6.
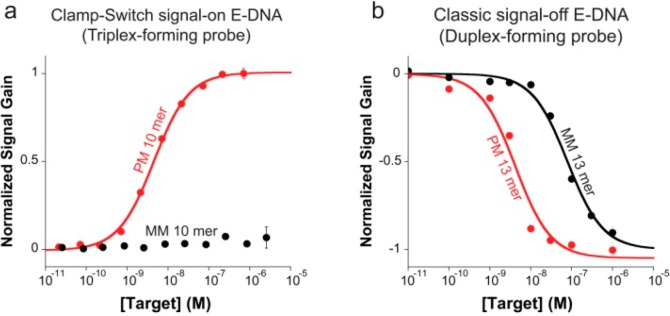
(a) Our E-DNA clamp-switch sensor is highly specific. We demonstrate this by interrogating the sensor with a perfect match and a one-base mismatch target (both 10-mer) at increasing concentrations. The affinity of the mismatch target is at least 2000-fold poorer than that of the perfect match target, thus demonstrating that the sequence-specific Hoogsteen base pairs in the clamp-switch offer an additional specificity check that increases the probe’s specificity compared to an equivalent E-DNA sensor based solely on Watson–Crick interactions. (b) As a further demonstration of this, we show here the binding curves obtained with a perfect match and a one-base mismatch using a classic E-DNA sensor based on a linear DNA probe. This sensor (signal-off) shows a separation between the perfect-match and mismatch affinity of only ∼20-fold. These binding curves were obtained by adding increasing concentration of a perfectly matched target and a one-base mismatch target (10-mer for the clamp-switch and 13-mer for the linear probe) in 2 mL of 10 mM TRIS buffer, 10 mM MgCl2, and 100 mM NaCl.
