Abstract
Ezrin, a protein belonging to the Ezrin, radixin and moesin (ERM) family, was engaged in the metastatic spread of osteosarcoma. The Protein 4.1, Ezrin, radixin, moesin (FERM) domain of Ezrin binds the membrane Phosphatydil inositol (4,5) bisphosphate (PIP2), a crucial molecule belonging to the Phosphoinositide (PI) signal transduction pathway. The cytoskeleton cross-linker function of Ezrin largely depends on membrane PIP2 levels, and thus upon the activity of related enzymes belonging to the PI-specific phospholipase C (PI-PLC) family. Based on the role of Ezrin in tumour progression and metastasis, we silenced the expression of Vil2 (OMIM *123900), the gene which codifies for Ezrin, in cultured human osteosarcoma 143B and Hs888 cell lines. After Ezrin silencing, the growth rate of both cell lines was significantly reduced and morphogical changes were observed. We also observed moderate variations both of selected PI-PLC enzymes within the cell and of expression of the corresponding PLC genes. In 143B cell line the transcription of PLCB1 decreased, of PLCG2 increased and of PLCE differed in a time-dependent manner. In Hs888, the expression of PLCB1 and of PLCD4 significantly increased, of PLCE moderately increased in a time dependent manner; the expression of PLCG2 was up-regulated. These observations indicate that Ezrin silencing affects the transcription of selected PLC genes, suggesting that Ezrin might influence the expression regulation of PI-PLC enzymes.
Keywords: Signal transduction, PLC, Ezrin, Cytoskeleton, Osteosarcoma, Metastasis
Introduction
Osteosarcoma, the most common primary bone tumour in childhood and adolescence, includes several pathological entities, differing in clinical, radiological, and histological features (Mirabello et al. 2009a, b; Gatta et al. 2005). The presence of metastasis confers worse prognosis to the clinical outcome of osteosarcoma affected patients (Meyers et al. 2005). The identification of molecules involved in the metastasizing process is crucial in order to understand the mechanisms of tumour dissemination, possibly opening the way to novel therapeutic strategies.
Ezrin, a protein involved in the metastatic spread of osteosarcoma (Khanna et al. 2004), belongs to the Ezrin-radixin-moesin (ERM) family proteins, which play structural and regulatory roles (Khanna et al. 2004; Ferrari et al. 2008; Hunter 2004). The reduction of Ezrin significantly reduced the metastatic dissemination in osteosarcoma animal models (Khanna et al. 2004). Despite a number of observations following great research efforts, the mechanisms by which Ezrin mediates the metastatic process remain to be fully delineated.
The Protein 4.1, Ezrin, radixin, moesin (FERM) domain (Chishti et al. 1998) present in Ezrin is involved in the recognition of Phosphatydil inositol (4,5) bisphosphate (PIP2), a crucial molecule belonging to the Phosphoinositide (PI) signal transduction pathway (Gautreau et al. 1999; Martin 2003; Pujuguet et al. 2003; Zhao et al. 2004; Tsukita and Yonemura 1997; Fievet et al. 2007). The role of Ezrin in actin assembly (Defacque et al. 2000, Defacque et al. 2002) largely depends on the membrane PIP2 levels (Hao et al. 2009). ERM proteins bind actin and, by means of their N-terminal domains, simultaneously the PIP2 located at the plasma membrane (Niggli, V et al. 2008, Gilmore and Burridge 1996 Isenberg and Niggli 1998, Nakamura et al. 1999, Eberle et al. 1990, Dobos et al. 1992, Apgar 1995, Gachet et al. 1997, Gratacap et al. 1998). Once activated, Ezrin, commonly localized in the cytoplasm in its inactive form, moves and tethers actin to the cortical membrane, thus promoting cytoskeleton reorganization and subsequent cell morphology alterations (Dard et al. 2004; Zhu et al. 2007; Di Cristofano et al. 2010; Yang et al. 2012; Zhao et al. 2011; Tan and Yang 2010). Beside phosphorylation, activation of ERM proteins also occurs after interaction with PIP2 (Gilmore and Burridge 1996, Hirao et al. 1996, Legg and Isacke 1998, Nakamura et al. 1999). The levels of PIP2 are regulated by means of the PI-specific Phospholipase C family of enzymes Berridge and Dupont (1994); Divecha and Irvine 1995; Hisatsune et al. 2005; Rhee 2001Bunney and Katan 2011). The reduction of PIP2 levels induces ERM protein dissociation from the membrane, and PI-PLC activity is required for this chemokine-mediated event (Brown et al. 2011).
In mammals, thirteen PI-PLC enzymes were divided into six sub-families on the basis of amino acid sequence, domain structure and mechanism of recruitment (Suh et al. 2008). Regulatory domains specific to each subfamily determine the susceptibility to different mechanisms of activation (Suh et al. 2008). The distribution of PI-PLC enzymes seems strictly tissue specific (Suh et al. 2008; Lo Vasco 2010; Lo Vasco 2012), and probably each cell type owns a specific panel of expression, which differs under different stimulation conditions (Suh et al. 2008, Lo Vasco et al. 2012, Lo Vasco et al. 2007a, Lo Vasco et al. 2010b; Lo Vasco et al. 2010c, Lo Vasco et al. 2007b).
In order to investigate the PI-PLC isoforms specifically involved in the PIP2-mediated regulation of Ezrin activity, we silenced the transcript of Vil2, the gene which codifies for Ezrin (OMIM *123900) by transfecting cultured human osteosarcoma cell lines, 143B and Hs888, with specific antisense silencing RNA (siRNA). We analysed the effects of Ezrin silencing upon the survival and morphology of the cells, upon the expression of PLC genes, which codify for PI-PLC enzymes, and upon the localization PI-PLC enzymes both in transfected and in control cells.
Materials and methods
Cell cultures
Two human osteosarcoma cell lines were analysed, 143B and Hs888, obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10–15 % foetal bovine serum (FBS), 1 mM sodium pyruvate, 100 U/mL of penicillin, and 100 mg/mL of streptomycin, at 37 °C and 5 % CO2 according to ATCC recommendations.
Cells were grown at 37 ºC in a humidified 5 % CO2 atmosphere in an incubator (Forma Scientific, USA). Confluent monolayer of cells was rinsed with phosphate-buffered saline (PBS) and 0.25 % Trypsin/EDTA (disodium ethylene diaminetetraacetate) was added for 3–5 min at 37 ºC, gently shaking the flask, then neutralised using growth medium. Cells were counted using a Neubauer haemocytometer (Weber Scientific International Ltd., Middlesex, UK). Cells were stored at −20 °C until use.
Cell survival Trypan Blue test
Cells were diluted 1:1 in trypan blue (Sigma Aldrich, Dorset, UK) for survival quantification. A growth curve was designed counting the quantity of cells by cm2 at different times. The number of viable cells was determined by adding 0.4 % Trypan blue staining to an equal volume of cell suspension. Viable cells were counted using a Neubauer haemocytometer and a phase contrast microscope. The following equation was used to calculate the total number of viable cells in 1 ml suspension: number of total viable cells in 1 ml (TC) = *2*10^4 (=average of the cell counts from the squares of the haemocytometer grid, 2 = dilution factor 1:1). The number of live cells was used to determine the growth rate and experiments were repeated three times.
Cells transfection for Ezrin silencing
143B and Hs888 cells were transiently transfected with Ezrin silencing RNA using METAFECTENE SI + (Biontex Laboratories GmbH, Munich, Germany). siRNA sequences targeting Ezrin and negative control siRNA, were designed and synthesized by Invitrogen (Life Technologies, Foster City, CA, USA). The siRNA was designed according to Ezrin complementary DNA (cDNA) sequence (EZR Gene ID: 7430). Briefly, 2.2 ml cell suspension were prepared in complete cell culture medium with a concentration of 1,5 · 10^5 cells/ml of 143B cells and 3 · 10^5 cells/ml of Hs888. Cells were seeded, in 6-well plates, shortly before the addition of the lipoplex, according to the manufacturer’s instructions. Then cells were incubated under normal culture conditions (37 °C in CO2–containing atmosphere) until the lipoplex addition. Before transfection, 150 μl of 1× SI + buffer were mixed with 72 μl of METAFECTENE® SI + and 540 pMol of RNA stock solution. The mixture was incubated for 15 min at room temperature and then added to the cells within one hour from seeding. Cells were incubated 72 h. Functional siRNA was measured by reverse transcription–polymerase chain reaction (RT-PCR) and western blot analysis 24, 48 and 72 h after transfection. Contemporarily, a growth curve was designed counting cells using a Neubauer haemocytometer.
RNA extraction
Total RNA was extracted with a SV Total RNA Isolation System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The cells were transferred to a microcentrifuge tube containing 175 μL of SV RNA Lysis Buffer and were passed through a 20-gauge needle to shear the genomic DNA for 4 to 5 times. 350 μL of SV Dilution Buffer was then added, mixed by inverting 4 times, and placed in a heating block at 70 °C for 3 min. The sample was centrifuged for 10 min at 14,000 × g. The lysate solution was transferred to a new microcentrifuge tube, 200 μL of 95 % ethanol were added, and the mixture was transferred to a spin column assembly, and centrifuged at 14,000 × g for one minute. The liquid was discarded from the collection tube, 600 μL of SV RNA Wash Solution was added to the column, centrifuged at 14,000 × g for one minute, and the collection tube was emptied. A DNase incubation mixture was prepared per sample by combining 40 μL Yellow Core Buffer (Promega), 5 μL 0.09 M MnCl2 and 5 μL of DNase I enzyme. The DNase incubation mixture was added directly to the membrane of the spin basket. The mixture was incubated for 15 min at room temperature, 200 μL of SV DNase Stop Solution was added to the spin basket, and centrifuged at 14,000 × g for one minute. Next, 600 μL of SV RNA Wash Solution was added and centrifuged at 14,000 x g for one minute. The collection tube was emptied, 250 μL of SV RNA Wash Solution was added, and centrifuged at 14,000 × g for two minutes. The spin basket was transferred from the collection tube to an elution tube, 100 μL of Nuclease-Free Water was added to the membrane and centrifuged at 14,000 x g for one minute. Finally, RNA was eluted into a sterile collection tube with RNase-free water. The procedure was carried out for each sample. The concentration and quality of the RNA obtained was monitored using a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Inc. USA).
Reverse Transcriptase (RT)-Polymerase Chain Reaction (PCR)
RNA was reverse-transcribed into cDNA using High-Capacity cDNA Reverse Transcription Kit (Life Technologies, Foster City, CA, USA). Briefly, 2 μg RNA were incubated with the master mix (2 μl of 10 × Reverse Transcription Buffer, 0.8 μl of 25 x dNTPs (100 mM), 2 μl of 10 x random primers, 1 μl of MultiScribeTM Reverse Transcriptase (50 U/μl) and 3.2 μl of DNase-free water). 10 μl of diluted RNA was then added to make a final volume of 20 μl. The RNA mix was then amplified for 10 min at 25 °C, 120 min at 37 °C and 5 min at 85 °C in a Gene Amp® PCR System 9700 (Applied Biosystems) thermocycler.
The primer pairs (Bio Basic Inc, Amherst, New York, USA) are listed in Table 1. To amplify glyceraldehyde 3 phosphate dehydrogenase (GAPDH) (Bio Basic Inc, Amherst, New York, USA) the following primer pair was used: forward 5′ -CGAGATCCCTCCAAAATCAA-3′ reverse 5′-GTCTTCTGGGTGGCAGTGAT-3′. The specificity of the primers was verified by searching in the NCBI database for possible homology to cDNAs of unrelated proteins. RNA samples were also amplified by PCR without RT to exclude possible contamination.
Table 1.
Primers’ pairs for Polymerase Chain Reaction
| PI-PLC β1 (PLCB1; OMIM *607120) | forward 5’-AGCTCTCAGAACAAGCCTCCAACA-3’ reverse 5’-ATCATCGTCGTCGTCACTTTCCGT-3’ |
| PI-PLC β2 (PLCB2; OMIM *604114) | forward 5’-AAGGTGAAGGCCTATCTGAGCCAA-3’ reverse 5’-TTGGCAAACTTCCCAAAGCGAGT-3’ |
| PI-PLC β3 (PLCB3; OMIM *600230) | forward 5’-TATCTTCTTGGACCTGCTGACCGT-3’ reverse 5’-TGTGCCCTCATCTGTAGTTGGCTT-3’ |
| PI-PLC β4 (PLCB4; OMIM *600810) | forward 5’-GCACAGCACACAAAGGAATGGTCA-3’ reverse 5’-CGCATTTCCTTGCTTTCCCTGTCA-3’ |
| PI-PLC γ1 (PLCG1; OMIM *172420) | forward 5’-TCTACCTGGAGGACCCTGTGAA-3’ reverse 5’-CCAGAAAGAGAG CGTGTAGTCG-3’ |
| PI-PLC γ2 (PLCG2; OMIM *600220) | forward 5’-AGTACATGCAGATGAATCACGC-3’ reverse 5’-ACCTGAATCCTGATTTGACTGC-3’ |
| PI-PLC δ1 (PLCD1; OMIM *602142) | forward 5’-CTGAGCGTGTGGTTCCAGC-3’ reverse 5’-CAGGCCCTCGGACTGGT-3’ |
| PI-PLC δ3 (PLCD3; OMIM *608795) | forward 5’-CCAGAACCACTCTCAGCATCCA-3’ reverse 5’-GCCA TTGTTGAGCACGTAGTCAG-3’ |
| PI-PLC δ4 (PLCD4; OMIM *605939) | forward 5’-AGACACGTCCCAGTCTGGAACC- 3’ reverse 5’-CTGCTTCCTCTTCCTCATATTC- 3’ |
| PI-PLC ε (PLCE; OMIM *608414) | forward 5’-GGGGCCACGGTCATCCAC-3’ reverse 5’-GGGCCTTCATACCGTCCATCCTC-3’ |
| PI-PLC η1 (PLCH1; OMIM *612835 | forward 5’-CTTTGGTTCGGTTCCTTGTGTGG-3’ reverse 5’-GGATGCTTCTGTCAGTCCTTCC-3’ |
| PIPLC η2 (PLCH2; OMIM *612836) | forward 5’-GAAACTGGCCTCCAAACACTGCCCGCCG-3’ reverse 5’-GTCTTGTTGGAGATGCACGTGCCCCTTGC-3’ |
Standard analytical PCR reaction was performed with GoTaq Master Mix (Promega). Each PCR tube contained the following reagents: 5X GoTaq buffer, 0.2 μM forward primer, 0.2 μM reverse primer, 0.2 mM dNTPs, 0.5 mM MgCl2, 1.25 U GoTaq and 3,5 μl of (about 35 μg) template cDNA at a 50 μl final volume. Cycling conditions were performed with 95 °C initial denaturation step for 1 min was followed by 40 cycles consisting of 95 °C denaturation (30s), annealing (30 s) at the appropriate temperature for each primer pair and 72 °C extension (1 min) in Gene Amp® PCR System 9700 (Applied Biosystems) thermocycler. Amplified PCR products were visualized by 1.5 % TAE ethidium bromide-stained agarose gel electrophoresis for 1 h at 100 V using UV light transilluminator PC-assisted CCD camera UVB lamp (Vilber Lourmaret, France) was used for gel documentation. Gel electrophoresis of the amplification products revealed single DNA bands with nucleotide lengths as expected for all primer pairs.
Real-time PCR
The messenger RNA (mRNA) transcription of transfected 143B and Hs888 cells and normal controls was measured. Gene expression was analysed by real-time PCR using the 7500 Real-Time PCR instrument from Applied Biosystems™. TaqMan® primers and probes for each gene, as well as the GAPDH reference gene, were obtained from Applied Biosystems™. Briefly, total RNA was extracted with a SV Total RNA Isolation System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The purity and quantity of RNA was assessed by NanoDrop ND–1,000 Spectrophotometer (Thermo Fisher Scientific, Inc. USA). The RNA was reverse transcribed into cDNA with High Capacity cDNA Reverse Transcription Kit (Life Technologies, Foster City, CA, USA).
Amplification products were detected using gene-specific primers and probes labelled with reporter day FAM which yielded a predicted amplicons of 82, 84, 61, 78, 64, 93 and 62 base pairs respectively; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal standard, which yielded a predicted amplicon of 58 base pairs. Reaction mixtures for all gene expression assays contained: 5 μl TaqMan® mastermix (2X; Applied Biosystems™), 0,5 μl primer/probe mix specific for each analysed gene and 1 μl PCR grade water. 3,5 μl of cDNA (35 ng) were added. PCR reaction was carried out in triplicate on 96-well plate with 10 uL per well using 1X TaqMan Master Mix. After 2 min incubation at 50 °C and 10 min at 95 °C, the reaction was carried out for 40 cycles at 95 °C for 15 s and 60 °C for 1 min. At the end of the reaction, the results were evaluated using the ABI PRISM 7500 software. The Ct (Cycle threshold) values for each set of three reactions were averaged for calculations. The 2^-∆∆Ct method was used to calculate relative changes in gene expression.
Western blot
Western blot analyses were conducted 24 and 48 h from transfection and in normal controls. Cells were washed with cold PBS, then were processed in cell lysis buffer (50 mM Tris–HCl, 150 mM NaCl, 2 mM EDTA, 1 % NP–40, 2 mM sodium fluoride, 0.5 % sodium deoxycholate, and 0.1 % SDS) containing protease inhibitors. 50ug of protein was separated by 10 % sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes (Invitrogen, Life Technologies, CA, USA). The membranes were blocked in PBS with 5 % skim milk for 1 h and incubated overnight with the primary antibodies. Finally, membranes were visualized by the addition of anti-mouse immunoglobulin G (Jackson Immunoresearch, UK) and anti-rabbit immunoglobulin G (Jackson Immunoresearch, UK) enhanced chemiluminescence. Expression of β-actin was used as an internal control to normalize results. The densities of the bands on the membrane were scanned and analysed with ImageJ software.
Immunofluorescence analysis of subcellular distribution of target molecules
Immunofluorescence detection of Ezrin, PI-PLC ε, PI-PLC β1, PI-PLC γ2, PI-PLC δ4 expression was performed on coverslips cultured transfected and non-trasfected cells. Cells were washed three times with PBS and fixed with 4 % paraformaldehyde (PFA) in phosphate buffer saline (PBS) for 10 min at 4 °C, followed by three washes with PBS. Cells were incubated with primary antibodies diluted in PBS for 1 h at room temperature. Cover-slips were then incubated with the specific secondary antibody Texas Red or fluorescein-conjugated for 1 h at room temperature. Cells were washed twice with 1X PBS 5 min, then counterstained with 4',6-diamidino-2-phenylindole (DAPI) fluorescent staining. The slides were visualized using an inverted microscope.
Statistical analysis
For in vitro studies, differences were determined either with two-way repeated measures analysis of variance (ANOVA) with Bonferroni's multiple comparison test, and by student's one tailed t-test, using Prism 5.0a software (GraphPad Software, San Diego, CA, USA). A p value <0.05 was considered significant.
Results
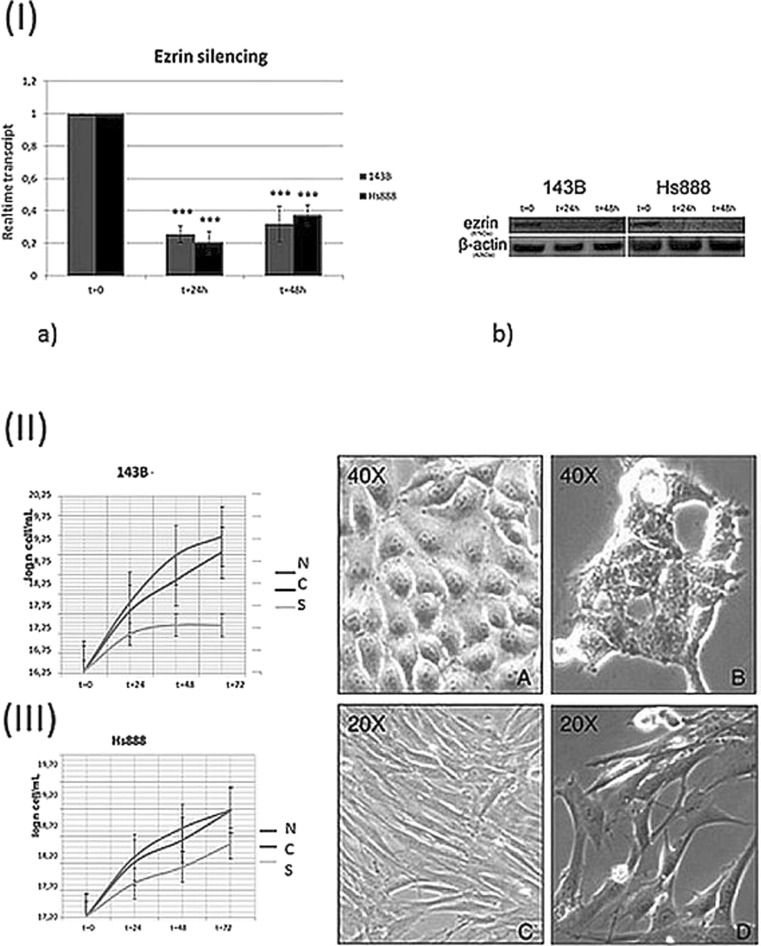
Silencing of Ezrin was validated by western blot, RT-PCR and gel electrophoresis, and real-time PCR of mRNA extracts and compared to non-targeting control siRNA (Fig. 1, I. Western blot assay showed no change in the expression of β-actin as internal control; the expression of Ezrin protein was significantly decreased in Ezrin siRNA transfected 143B cells compared to the transfected cells (Fig. 1, I b). Ezrin transcription was compared in cells transfected with Ezrin-silencing specific siRNA to control group, comprising untransfected cells and cells transfected with the carrier metefectamine. The transcription of Ezrin was suppressed in transfected 143B compared to the control group (not transfected and siRNA transfected), which correctly expressed Ezrin mRNA. The mRNA expression level of Ezrin in transfected cells was significantly reduced with respect to untreated cells (p < 0,001) (Fig. 1, a).
Fig. 1.
Effectiveness of Ezrin silencing. a Istogram of mRNA transcript concentrations after 0, 24 and 48 h from Ezrin silencing in 143B (gray) and Hs888 (black) cell lines. b Gel electrophoresis from Western blot of Ezrin protein in 143B and Hs888 cell lines compared to actin protein loading control. Growth curve after Ezrin silencing (left) effect of Ezrin siRNA on cell morphology (right) in 143B cells: the growth of silenced cells is significantly slowed with respect to untreated cells (p < 0,5). Bar errors are indicated. N = untrasfected control 143B cells; C = metafectamine transfected control 143B cells; Ezrin siRNA transfected 143B cells. II a and b - morphological changes in 143B cells (40X, contrast-phase microscopy). Irregular outline of the plasma membrane, reduced intercellular adhesion and cytoplasm micro-vacuolization. Growth curve after Ezrin silencing (left) effect of Ezrin siRNA on cell morphology (right) in Hs888 cells: the growth of silenced cells is slowed with respect to untreated cells. Bar errors are indicated. N = untrasfected control Hs888 cells; C = metafectamine transfected control Hs888 cells; Ezrin siRNA transfectedHs888 cells. III c and d - morphological changes in Hs888 cells (20, contrast-phase microscopy). Quantitative reduction of cells displaying substantially well-preserved structure
In 143B Cells, survival Trypan Blue test indicated decrease of the growth rate of Ezrin siRNA-transfected cells (Fig. 1, II) with respect to control cells (p < 0,5). The growth rate was reduced in Ezrin siRNA transfected cells (S) in the 0–72 h interval with respect to untrasfected control cells (N) and in metafectamine transfected control cells (C). The cell growth was reduced in 143B cells in which Ezrin was silenced with respect to controls since 3–6 h from silencing. The growth rate of S cells was reduced after 24 h from Ezrin silencing (Fig. 1, II), and remained constant during the remaining 24–72 h interval. The growth rate of N and C cells had a significant exponential progression after 24 h. In 143B transfected cells the expression of PLCG2 gene increased about 40 % after 24 h from the transfection and about 25 % after 48 h; the expression of PLCB1 decreased about 55 % (Fig. 2). The expression of PLCE was moderately (about 15 %) reduced after 24 h from the silencing and moderately increased after 48 h (Fig. 2). PLCB1 was expressed in control cells in low concentration. PLCD4, undetected in 143B, increased about two folds after 24 h from Ezrin silencing (Fig. 2). There was a statistically significant difference in the mRNA expression levels of PLCB1, which was decreased both after 24 (p < 0,0005) and 48 (p < 0,0125) hours from Ezrin silencing. The expression of GAPDH mRNA in the considered interval did not differ in a statistically significant manner, as expected.
Fig. 2.
Real-time results after Ezrin silencing. Istograms of the transcript concentrations of PLC genes after 0, 24 and 48 h from Ezrin silencing in 143B (gray) and Hs888 (black) cell lines
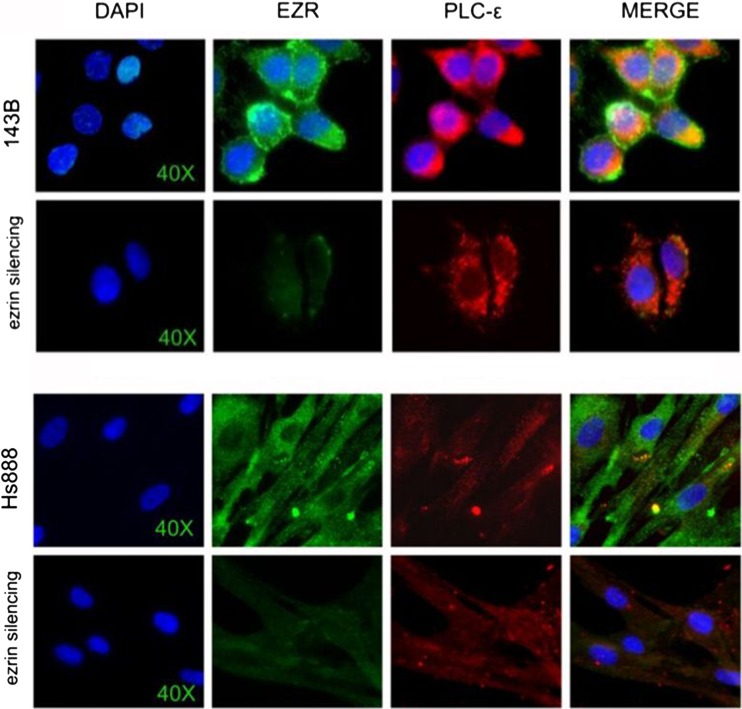
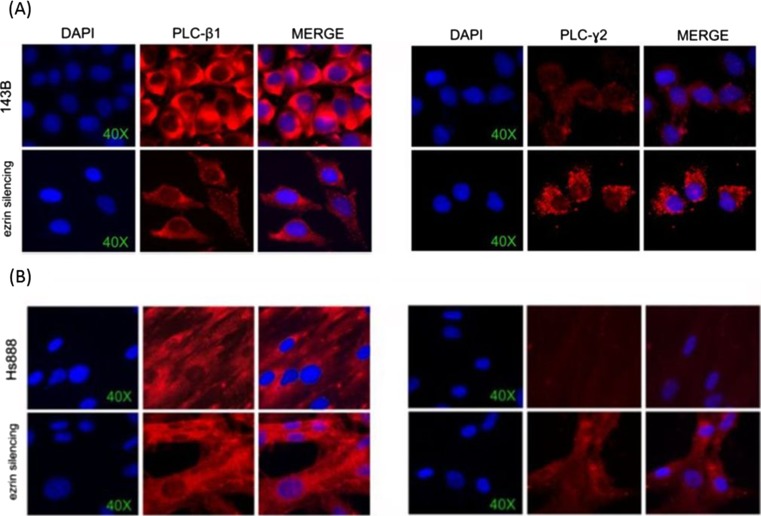
Immunofluorescence microscopy showed moderate signal intensity of Ezrin localized in the cytoplasm, with membrane signal enhancement in 143B cell controls. PI-PLC ε was also localized in the cytoplasm, with weak signal intensity. A focal cytoplasmic co-localization of Ezrin and PI-PLC ε was observed (Fig. 3). In the cytoplasm, PI-PLC β1 and PI-PLC γ2 were respectively strongly and weakly detected (Fig. 4). In 143B cells transfected with Ezrin siRNA, irregular outline of the plasma membrane was associated with reduced intercellular adhesion and micro-vacuolization of the cytoplasm was also observed at optic microscope (Fig. 1, II A and B). Significant reduction of the signal intensity of PI-PLC β1 in the cytoplasm was also detected (Fig. 4). For PI-PLC γ2 stronger cytoplasmic signal with membrane staining reinforcement were observed (Fig. 4). Moderate increase and of PI-PLC ε signal intensity, localized in cytoplasm was observed (Fig. 3).
Fig. 3.
Partial co-localization of Ezrin and PI-PLC ε Fluorescence immunocytochemistry of Ezrin (green) and PI-PLC ε (red) in 143B (upper) and Hs888 (lower) cell lines. Diaminophenyl indole (DAPI, blue) counterstain for nuclei (60X)
Fig. 4.
Immunofluorescence analyses. a Localization of PI-PLC β1 and PI-PLC γ2 in 143B cell line. LEFT. Fluorescence immunocytochemistry of PI-PLC β1 (red) in 143B (upper) and in 143B cells after Ezrin silencing (lower). Diaminophenyl indole (DAPI, blue) counterstain for nuclei (60X). RIGHT. Fluorescence immunocytochemistry of PI-PLC γ2 (red) in 143B (upper) and in 143B cells after Ezrin silencing (lower). Diaminophenyl indole (DAPI, blue) counterstain for nuclei (60X). b Localization of PI-PLC β1 and PI-PLC γ2 in Hs888 cell line LEFT. Fluorescence immunocytochemistry of PI-PLC β1 (red) in Hs888 (upper) and in Hs888 cells after Ezrin silencing (lower). Diaminophenyl indole (DAPI, blue) counterstain for nuclei (60X). RIGHT. Fluorescence immunocytochemistry of PI-PLC γ2 (red) in Hs888 (upper) and in Hs888 cells after Ezrin silencing (lower). Diaminophenyl indole (DAPI, blue) counterstain for nuclei (60X)
In Hs888 Cells, survival Trypan Blue test indicated that the growth rate of the Ezrin siRNA-treated cells decreased in a time-dependent manner (Fig. 1) with respect to control cells (p < 0.5). The growth rate was reduced in Ezrin siRNA transfected cells (S) in the 0–24 h interval with respect to untrasfected control cells (N) and in metafectamine transfected control cells (C) (Fig. 1, III).
The expression of PLCE was comparable to the untreated cells after 24 h, and increased about 80 % after 48 h (Fig. 2); PLCG2 was moderately increased; PLCB1, weakly expressed in untreated controls, significantly increased (from 12 to 16 folds) in the 24–48 h interval (Fig. 2); PLCD4 increased after 24 h from silencing (Fig. 2). A statistically significant difference in the mRNA expression levels of PLCE (p < 0,005) was calculated comparing Ezrin siRNA transfected Hs888 and control cells (both untrasfected and metafectamine transfected cells). After 48 h from Ezrin silencing a statistically significant difference of mRNA expression levels of PLCB1 both after 24 (p < 0,0005) and 48 (p < 0,0125) hours from transfection was also calculated. The expression of GAPDH mRNA in the considered interval did not differ in a statistically significant manner, as expected. Immunofluorescence microscopy showed focal cytoplasmic co-localization of Ezrin and PI-PLC ε in Hs888 control cells (Fig. 3). After Ezrin silencing, the quantitative reduction of cellular elements was accompanied by substantially well-preserved structure (Fig. 1, c and d). Ezrin was mildly localized in the cytoplasm. PI-PLC ε was weakly localized in the cytoplasm (Fig. 3). PI-PLC β1 was localized in the cytoplasm in control cells, and Ezrin silencing induced a significant increase of the signal intensity (Fig. 4). Moderate signal intensity for PI-PLC γ2 was detected in the cytoplasm, with strong perinuclear enhancement (Fig. 4). After Ezrin silencing, a slight increase of PI-PLC ε signal intensity was observed, mainly localized in the cytoplasm (Fig. 3).
Discussion
The present results in 143B cells and Hs888 are not comparable, probably due to the different origins of the cells. 143B thymidine kinase negative human osteosarcoma cells, originating from highly aggressive primary tumour, develop osteolytic tumours (Kaminski et al. 2003). Hs888 cells derived from lung metastasis of osteosarcoma. Ezrin silencing induced cell growth rate reduction more marked in 143B than in Hs888 cells.
Ezrin silencing reduced the growth rate of cells. That was more marked in 143B line, as well as morphological changes, probably related to the cytoskeleton-linker activity of Ezrin, and microvacuolization of the cytoplasm. The quantitative changes of PI-PLC enzymes occurring after 24 h from Ezrin silencing might indicate that lack of Ezrin affects the regulation of the PI signal transduction pathway. The changes of the quantity and localization of PI-PLC γ2 corroborate our previous hypothesis that this isoform might play an important role in osteosarcoma.
The differences of intracellular PI-PLC enzymes accorded to the expression of the corresponding PLC genes (Fig. 2). In both cell lines, basal PI-PLC β1 is mildly expressed (2–4.5 ng/ml), according to previous observations (Lo Vasco et al. 2014). PI-PLC β1 is selectively increased during myoblast and adipocyte differentiation (Faenza et al. 2004, O'Carroll et al. 2009), and might be altered in breast cancer (Molinari et al. 2012, Abalsamo et al. 2012). Evidences suggested that deletion of PLCB1 favours cancer progression in the myeloid lineage (Lo Vasco et al. 2004, Kaminskas et al. 2005) and is involved in differentiation. Therefore, this isoform might contrast cancer progression. In Hs888, Ezrin silencing induced a very significant increase of PLCB1 transcription (Fig. 2) and of cytoplasmic PI-PLC β1 (Fig. 4). The hypothesis that, in the metastatic Hs888 cell line, the expression of PI-PLC β1 might be under the control of Ezrin will require studies in order to investigate whether this mechanism occurs directly or involves further signalling molecules.
Ezrin silencing induced PLCG2 transcription and cytoplasmic PI-PLC γ2 increase. The PI-PLC γ enzymes, detected at higher level in tumour than in normal tissues (Arteaga et al. 1991, Noh et al. 1995), are characterized by a unique region comprising two tandem SH2 and one SH3 domains (Katan and Williams 1997, Bunney and Katan 2011). Ezrin can interact with the SH2 domain, and might act as negative regulator of PI-PLC γ2. The up-regulation of PI-PLC γ2 following Ezrin silencing might accord to the osteolytic nature of 143B cells (Kaminski et al. 2003). In fact, PI-PLC γ2 is involved in actin cytoskeleton reorganization (Cremasco V. 2999), and, in osteoclasts, is required for early phase differentiation (Kertész et al. 2012), adhesion, migration, bone resorption (Epple et al. 2008), regulation of the Proto-oncogene Src activation and membrane localization Mao et al. (2006) PI-PLC γ2 was also indicated as a critical regulator in bone and immune cells during autoimmune inflammation (Faccio and Cremasco 2010). On the other hand, PI-PLC γ2, usually absent in Hs888 cells, was detected in low concentration after Ezrin silencing. That suggests that PI-PLC γ2 might crucially network Ezrin. That observation might deserve great attention, as increasing evidences suggest that PI-PLC γ isoforms play key roles in cell migration and invasion (Lattanzio et al. 2013), as well as in cell growth and survival Mirabello et al. (2009a).
PI-PLC δ4, exclusively expressed in Hs888 cell line, accordingly to previous findings (Lo Vasco et al. 2013a), was up-regulated after Ezrin silencing. PI-PLC δ enzymes, the most primitive and evolutionary conserved, are very sensitive to calcium and might play a key role in cell proliferation (Suh et al. 2008; Liu et al. 1996; Fukami et al. 2000). In fact, PI-PLC δ4 is expressed more abundantly in high-rate proliferating cells (Santi et al. 2003, Ochocka and Pawelczyk 2003) and was associated to astrocytoma (Lo Vasco et al. 2007a, 2010a), and breast cancer (Leung et al. 2004). . In the present experiments, the increase of PLCD4 transcription might be related to the induced reduction of Ezrin, although the mechanism and the contemporary up-regulation of PLCB1 will require further studies.
In both cell lines, the transcription of PLCE was affected by Ezrin silencing in a time-related manner. 24 h after Ezrin silencing PLCE transcription was reduced and after 48 h increased, more markedly in Hs888 than in 143B cells. PI-PLC ε enzyme is thought to play an important role in carcinogenesis. However, the mechanism of action is not completely understood, and controversial data were reported. A number of evidences indicate that PI-PLC ε might favour cancer initiation and/or progression, as in bladder (Cheng et al. 2011; Ou et al. 2010), murine skin (Bai et al. 2004, Oka 2010, Li et al. 2009), head and neck cancers (Bourguignon et al. 2006). The rs 2274223 polymorphism was significantly associated to increased risk of squamous cell carcinoma and gastric cancer (Hao 2013 By contrast, tumour suppressive role for PI-PLC ε was recently suggested in Ras-triggered cancers (Martins et al. 2014). Our present results suggest the existence of a relationship between Ezrin and PI-PLC ε, corroborated by microscopy observations detecting focal and partial cytoplasmic co-localization (Figure 3).
Resuming, in the analysed osteosarcoma cell lines, Ezrin silencing reduced the growth rate and induced morphology changes, corroborating the hypothesis that it is involved in cell growth/survival and in cytoskeleton organization. The tight regulation of membrane PIP2 levels might represent a mechanism of control of Ezrin activity, directly under the control of the PI signal transduction system by means of activated PI-PLC enzymes. Both Ezrin and PI-PLC enzymes bind PIP2 in a competitive manner or, probably, in a more complex mechanism. Ezrin reduction or absence, induced by silencing the transcript of Vil2, increased the available PIP2 that might explain the observed re-modulation of the PI-PLC enzymes panel. That suggests a possible connection in terms of reciprocal regulation that will require further studies. The present data might also partially contribute to explain the role of the PI-PLC family in the cytoskeleton organization, which might be related to the actin cross-linker role of Ezrin. Further studies, addressed to elucidate the relationship between Ezrin and cell line-specific PI-PLC enzymes, might help to identify the crosstalk among the molecules, opening the way to novel insights in the progression of the disease, with special regard to metastatic spread inputs, and, as far as one can see, to novel therapeutic strategies.
Acknowledgments
The authors thank the‘Serena Talarico Association’ for supporting this research and precious encouragement.
References
- Abalsamo L, Spadaro F, Bozzuto G, Paris L, Cecchetti S, Lugini L, Iorio E, Molinari A, Ramoni C, Podo F. Inhibition of phosphatidylcholine-specific phospholipase C results in loss of mesenchymal traits in metastatic breast cancer cells. Breast Cancer Res. 2012;14(2):R50. doi: 10.1186/bcr3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apgar JR. Activation of protein kinase C in rat basophilic leukemia cells stimulates increased production of phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate: correlation with actin polymerization. Mol Biol Cell. 1995;6(1):97–108. doi: 10.1091/mbc.6.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga CL, Johnson MD, Todderud G, Coffey RJ, Carpenter G, Page DL. Elevated content of the tyrosine kinase substrate phospholipase C-gamma 1 in primary human breast carcinomas. Proc Natl Acad Sci U S A. 1991;88(23):10435–10439. doi: 10.1073/pnas.88.23.10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Edamatsu H, Maeda S, Saito H, Suzuki N, et al. Crucial role of phospholipase Ce in chemical carcinogen-induced skin tumor development. Cancer Res. 2004;64:8808–8810. doi: 10.1158/0008-5472.CAN-04-3143. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Dupont G. Spatial and temporal signalling by calcium. Curr Opin Cell Biol. 1994;6(2):267–274. doi: 10.1016/0955-0674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- Bourguignon LY, Gilad E, Brightman A, Diedrich F, Singleton P. Hyaluronan-CD44 interaction with leukemia-associated RhoGEF and epidermal growth factor receptor promotes Rho/Ras co-activation, phospholipase C epsilon-Ca2+ signaling, and cytoskeleton modification in head and neck squamous cell carcinoma cells. J Biol Chem. 2006;281(20):14026–14040. doi: 10.1074/jbc.M507734200. [DOI] [PubMed] [Google Scholar]
- Brown JB, Cheresh P, Goretsky T, Managlia E, Grimm GR, Ryu H, Zadeh M, Dirisina R, Barrett TA. Epithelial phosphatidylinositol-3-kinase signaling is required for β-catenin activation and host defense against Citrobacter rodentium infection. Infect Immun. 2011;79(5):1863–1872. doi: 10.1128/IAI.01025-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunney TD, Katan M. PLC regulation: emerging pictures for molecular mechanisms. Trends Biochem Sci. 2011;36(2):88–96. doi: 10.1016/j.tibs.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Cheng H, Luo C, Wu X, Zhang Y, He Y, et al (2011) shRNA Targeting PLCe1 Inhibits Bladder Cancer Cell Growth In Vitro and In Vivo. Urology 78: 474.e477–474.e411 [DOI] [PubMed]
- Chishti AH, Kim AC, Marfatia SM, Lutchman M, Hanspal M, Jindal H, Liu SC, Low PS, Rouleau GA, Mohandas N, Chasis JA, Conboy JG, Gascard P, Takakuwa Y, Huang SC, Benz EJ, Jr, Bretscher A, Fehon RG, Gusella JF, Ramesh V, Solomon F, Marchesi VT, Tsukita S, Tsukita S, Hoover KB, et al. The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem Sci. 1998;23(8):281–282. doi: 10.1016/S0968-0004(98)01237-7. [DOI] [PubMed] [Google Scholar]
- Cremasco V., Benasciutti E., Cella M., Kisseleva M., Croke M., Faccio R. Phospholipase C Gamma 2 Is Critical for Development of a Murine Model of Inflammatory Arthritis by Affecting Actin Dynamics in Dendritic Cells [DOI] [PMC free article] [PubMed]
- Dard N, Louvet-Vallee S, Santa-Maria A, et al. Phosphorylation of Ezrin on threonine T567 plays a crucial role during compaction in the mouse early embryo. Dev Biol. 2004;271:87–97. doi: 10.1016/j.ydbio.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Defacque H, Egeberg M, Habermann A, Diakonova M, Roy C, Mangeat P, Voelter W, Marriott G, Pfannstiel J, Faulstich H, Griffiths G. Involvement of Ezrin/moesin in de novo actin assembly on phagosomal membranes. EMBO J. 2000;19(2):199–212. doi: 10.1093/emboj/19.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defacque H, Bos E, Garvalov B, Barret C, Roy C, Mangeat P, Shin HW, Rybin V, Griffiths G. Phosphoinositides regulate membrane-dependent actin assembly by latex bead phagosomes. Mol Biol Cell. 2002;13(4):1190–1202. doi: 10.1091/mbc.01-06-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofano C, Leopizzi M, Miraglia A, Sardella B, Moretti V, Ferrara A, Petrozza V, Della RC. Phosphorylated Ezrin is located in the nucleus of the osteosarcoma cell. Mod Pathol. 2010;23(7):1012–1020. doi: 10.1038/modpathol.2010.77. [DOI] [PubMed] [Google Scholar]
- Divecha N, Irvine RF. Phospholipid signaling. Cell. 1995;80(2):269–278. doi: 10.1016/0092-8674(95)90409-3. [DOI] [PubMed] [Google Scholar]
- Dobos GJ, Norgauer J, Eberle M, Schollmeyer PJ, Traynor-Kaplan AE. C5a reduces formyl peptide-induced actin polymerization and phosphatidylinositol(3,4,5)trisphosphate formation, but not phosphatidylinositol (4,5) bisphosphate hydrolysis and superoxide production, in human neutrophils. J Immunol. 1992;149(2):609–614. [PubMed] [Google Scholar]
- Eberle M, Traynor-Kaplan AE, Sklar LA, Norgauer J. Is there a relationship between phosphatidylinositol trisphosphate and F-actin polymerization in human neutrophils? J Biol Chem. 1990;265(28):16725–16728. [PubMed] [Google Scholar]
- Epple H, Cremasco V, Zhang K, Mao D, Longmore GD, et al. Phospholipase Cgamma2 modulates integrin signaling in the osteoclast by affecting the localization and activation of Src kinase. Mol Cell Biol. 2008;28:3610–3622. doi: 10.1128/MCB.00259-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faccio R, Cremasco V. PLCgamma2: where bone and immune cells find their common ground. Ann N Y Acad Sci. 2010;1192:124–130. doi: 10.1111/j.1749-6632.2009.05217.x. [DOI] [PubMed] [Google Scholar]
- Faenza I, Bavelloni A, Fiume R, Santi P, Martelli AM, Billi AM, Lo Vasco VR, Manzoli L, Cocco L. Expression of phospholipase C β family isoenzymes in C2C12 myoblasts during terminal differentiation. J Cell Physiol. 2004;200(2):291–296. doi: 10.1002/jcp.20001. [DOI] [PubMed] [Google Scholar]
- Ferrari S, Zanella L, Alberghini M, et al. Prognostic significance of immunohistochemical expression of Ezrin in non-metastatic high-grade osteosarcoma. Pediatr Blood Cancer. 2008;50:752–756. doi: 10.1002/pbc.21360. [DOI] [PubMed] [Google Scholar]
- Fievet B, Louvard D, Arpin M. ERM proteins in epithelial cell organization and functions. Biochim Biophys Acta. 2007;1773:653–660. doi: 10.1016/j.bbamcr.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Fukami K, Takenaka K, Nagano K, Takenawa T. Growth factor-induced promoter activation of murine phospholipase Cδ4 gene. Eur J Biochem. 2000;267:28–36. doi: 10.1046/j.1432-1327.2000.00943.x. [DOI] [PubMed] [Google Scholar]
- Gachet C, Payrastre B, Guinebault C, Trumel C, Ohlmann P, Mauco G, Cazenave JP, Plantavid M, Chap H. Reversible translocation of phosphoinositide 3-kinase to the cytoskeleton of ADP-aggregated human platelets occurs independently of Rho A and without synthesis of phosphatidylinositol (3,4)-bisphosphate. J Biol Chem. 1997;272(8):4850–4854. doi: 10.1074/jbc.272.8.4850. [DOI] [PubMed] [Google Scholar]
- Gatta G, Capocaccia R, Stiller C, Kaatsch P, Berrino F, Terenziani M. EUROCARE Working Group Childhood cancer survival trends in Europe: a EUROCARE Working Group study. J Clin Oncol. 2005;3(16):3742–3751. doi: 10.1200/JCO.2005.00.554. [DOI] [PubMed] [Google Scholar]
- Gautreau A, Poullet P, Louvard D. Ezrin, a plasma membrane-microfilament linker, signals cell survival through the phosphatidylinositol 3-kinase/Akt pathway. Proc Natl Acad Sci U S A. 1999;96:7300–7305. doi: 10.1073/pnas.96.13.7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore AP, Burridge K. Molecular mechanisms for focal adhesion assembly through regulation of protein-protein interactions. Structure. 1996;4(6):647–651. doi: 10.1016/S0969-2126(96)00069-X. [DOI] [PubMed] [Google Scholar]
- Gratacap MP, Payrastre B, Viala C, Mauco G, Plantavid M, Chap H. Phosphatidylinositol 3,4,5-trisphosphate- dependent stimulation of phospholipase C-gamma2 is an early key event in FcgammaRIIA-mediated activation of human plate- lets. J Biol Chem. 1998;273(38):24314–24321. doi: 10.1074/jbc.273.38.24314. [DOI] [PubMed] [Google Scholar]
- Hao N-B, Ya-Fei HE, Zhang D, Luo G, Chen B-J, Zang Y, Yang S-M (2013) PLCE1 Polymorphism and Upper Gastrointestinal Cancer Risk: A Meta-Analysis. PLoS ONE 8(6):e67229 [DOI] [PMC free article] [PubMed]
- Hao JJ, Liu Y, Kruhlak M, Debell KE, Rellahan BL, Shaw S. Phospholipase C-mediated hydrolysis of PIP2 releases ERM proteins from lymphocyte membrane. J Cell Biol. 2009;184(3):451–462. doi: 10.1083/jcb.200807047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao M, Sato N, Kondo T, Yonemura S, Monden M, Sasaki T, Takai Y, Tsukita S, Tsukita S. Regulation mechanism of ERM (Ezrin/radixin/moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J Cell Biol. 1996;135(1):37–51. doi: 10.1083/jcb.135.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisatsune C, Nakamura K, Kuroda Y et al (2005) Amplification of Ca2+ signaling by diacylglycerolmediatedinositol 1,4,5-trisphosphate production. J Biol Chem 280(12):11723–30 [DOI] [PubMed]
- Hunter KW. Ezrin, a key component in tumor metastasis. Trends Mol Med. 2004;10:201–204. doi: 10.1016/j.molmed.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Isenberg G, Niggli V (1998) Interaction of cytoskeletal proteins with membrane lipids. Int Rev Cytol 178:73–125 [DOI] [PubMed]
- Kaminskas E, Farrell A, Abraham S, Baird A, Hsieh LS, Lee SL, Leighton JK, Patel H, Rahman A, Sridhara R, Wang YC (2005) Pazdur R; FDA approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res 11(10):3604–08 [DOI] [PubMed]
- Kaminski M, Masaoka M, Karbowski M, Kedzior J, Nishizawa Y, Usukura J, Wakabayashi T. Ultrastructural basis for the transition of cell death mode from apoptosis to necrosis in menadione-treated osteosarcoma 143B cells. J Electron Microsc. 2003;52:313–325. doi: 10.1093/jmicro/52.3.313. [DOI] [PubMed] [Google Scholar]
- Katan M, Williams RL. Phosphoinositide-specific phospholipase C: structural basis for catalysis and regulatory interactions. Semin Cell Dev Biol. 1997;8(3):287–296. doi: 10.1006/scdb.1997.0150. [DOI] [PubMed] [Google Scholar]
- Kertész Z, Gyori D, Körmendi S, Fekete T, Kis-Tóth K, Jakus Z, Schett G, Rajnavölgyi E, Dobó-Nagy C, Mócsai A. Phospholipase Cγ2 is required for basal but not oestrogen deficiency-induced bone resorption. Eur J Clin Invest. 2012;42(1):49–60. doi: 10.1111/j.1365-2362.2011.02556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna C, Wan X, Bose S, et al. The membrane-cytoskeleton linker Ezrin is necessary for osteosarcoma metastasis. Nat Med. 2004;10:182–186. doi: 10.1038/nm982. [DOI] [PubMed] [Google Scholar]
- Lattanzio R, Piantelli M, Falasca M (2013) Role of phospholipase C in cell invasion and metastasis. Adv Biol Regul 53(3):309–18. doi:10.1016/j.jbior.2013.07.006 [DOI] [PubMed]
- Legg JW, Isacke CM. Identification and functional analysis of the Ezrin-binding site in the hyaluronan receptor, CD44. Curr Biol. 1998;8(12):705–708. doi: 10.1016/S0960-9822(98)70277-5. [DOI] [PubMed] [Google Scholar]
- Leung DW, Tompkins C, Brewer J, Ball A, Coon M, Morris V, Waggoner D, Singer JW. Phospholipase C delta-4 over- expression upregulates ErbB1/2 expression, Erk signaling path- way, and proliferation in MCF-7 cells. Mol Cancer. 2004;3:15. doi: 10.1186/1476-4598-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Edamatsu H, Kitazawa R, Kitazawa S, Kataoka T. Phospholipase cepsilon promotes intestinal tumorigenesis of Apc(Min/+) mice through augmentation of inflammation and angiogenesis. Carcinogenesis. 2009;30(8):1424–1432. doi: 10.1093/carcin/bgp125. [DOI] [PubMed] [Google Scholar]
- Liu N, Fukami K, Yu H, Takenawa T. A new phospho- lipase Cδ4 is induced at S-phase of the cell cycle and appears in the nucleus. J Biol Chem. 1996;271:355–360. doi: 10.1074/jbc.271.1.355. [DOI] [PubMed] [Google Scholar]
- Lo VVR, Calabrese G, Manzoli L, Palka GD, Spadano A, Morizio E, Guanciali-Franchi P, Fantasia D, Cocco L. Inositide-specific Phospholipase C β1 gene deletion in the progression of Myelodisplastic Syndrome to Acute Myeloid. Leukemia. 2004;18(6):1122–1126. doi: 10.1038/sj.leu.2403368. [DOI] [PubMed] [Google Scholar]
- Lo Vasco VR. Signaling in the genomic era. J Cell CommunSignal. 2010;4(3):115–117. doi: 10.1007/s12079-010-0091-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Vasco VR. The Phosphoinositide pathway and the signal transduction network in neural development. Neurosci Bull. 2012;28(6):789–800. doi: 10.1007/s12264-012-1283-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Vasco VR, Fabrizi C, Artico M, Cocco L, Billi AM, Fumagalli L, Manzoli FA. Expression of phosphoinositide-specific phospholipase C isoenzymes in cultured astrocytes. J Cell Biochem. 2007;100(4):952–959. doi: 10.1002/jcb.21048. [DOI] [PubMed] [Google Scholar]
- Lo Vasco VR, Fabrizi C, Artico M, Cocco L, Billi AM, Fumagalli L, Manzoli FA. Expression of phosphoinositide-specific phospholipase C isoenzymes in cultured astrocytes. J Cell Biochem. 2007;100(4):952–959. doi: 10.1002/jcb.21048. [DOI] [PubMed] [Google Scholar]
- Lo Vasco VR, Fabrizi C, Panetta B, Fumagalli L, Cocco L. Expression pattern and sub cellular distribution of phosphoinositide specific phospholipase C enzymes after treatment with U-73122 in rat Astrocytoma cells. J Cell Biochem. 2010;110(4):1005–1012. doi: 10.1002/jcb.22614. [DOI] [PubMed] [Google Scholar]
- Lo Vasco VR, Fabrizi C, Fumagalli L, Cocco L. Expression of phosphoinositide specific phospholipase C isoenzymes in cultured astrocytes activated after stimulation with Lipopolysaccharide. J Cell Biochem. 2010;109(5):1006–1012. doi: 10.1002/jcb.22480. [DOI] [PubMed] [Google Scholar]
- Lo Vasco VR, Fabrizi C, Panetta B, Fumagalli L, Cocco L. Expression pattern and sub cellular distribution of phosphoinositide specific phospholipase C enzymes after treatment with U-73122 in rat Astrocytoma cells. J Cell Biochem. 2010;110(4):1005–1012. doi: 10.1002/jcb.22614. [DOI] [PubMed] [Google Scholar]
- Lo Vasco VR, Leopizzi M, Chiappetta C, Businaro R, Polonia P, Della Rocca C, Litta P. Expression of phosphoinositide-specific phospholipase C enzymes in normal endometrium and in endometriosis. Fertil Steril. 2012;98(2):410–414. doi: 10.1016/j.fertnstert.2012.04.020. [DOI] [PubMed] [Google Scholar]
- Lo Vasco VR, Leopizzi M, Chiappetta C, Puggioni C, Di Cristofano C, Della RC. Expression of Phosphoinositide-specific phospholipase C enzymes in human osteosarcoma cell lines. J Cell Commun Signal. 2013;7(2):141–150. doi: 10.1007/s12079-013-0194-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Vasco VR, Leopizzi M, Chiappetta C, Puggioni C, Della Rocca C, Businaro R. Lypopolysaccharide down-regulates the expression of selected phospholipase C genes in cultured endothelial cells. Inflammation. 2013;36(4):862–868. doi: 10.1007/s10753-013-9613-3. [DOI] [PubMed] [Google Scholar]
- Lo Vasco VR, Leopizzi M, Puggioni C, Della Rocca C, Businaro R. Fibroblast growth factor acts upon the transcription of phospholipase C genes in human umbilical vein endothelial cells. Mol Cell Biochem. 2014;388(1):51–59. doi: 10.1007/s11010-013-1898-x. [DOI] [PubMed] [Google Scholar]
- Mao D, Epple H, Uthgenannt B, Novack DV, Faccio R. PLCgamma2 regulates osteoclastogenesis via its interaction with ITAM proteins and GAB2. J Clin Invest. 2006;116(11):2869–2879. doi: 10.1172/JCI28775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GS. Cell signaling and cancer. Cancer Cell. 2003;4:167–174. doi: 10.1016/S1535-6108(03)00216-2. [DOI] [PubMed] [Google Scholar]
- Martins M, Mc Carthy A, Baxendale R, Guichard S, Magno L, Kessaris N, El-Bahrawy M, Yu P, Katan M. Tumor suppressor role of phospholipase Cε in Ras-triggered cancers. PNAS, e-pub January 31 2014, DOI:10.1073/pnas.1311500111 [DOI] [PMC free article] [PubMed]
- Meyers PA, Schwartz CL, Krailo M, Kleinerman ES, Betcher D, Bernstein ML, Conrad E, Ferguson W, Gebhardt M, Goorin AM, Harris MB, Healey J, Huvos A, Link M, Montebello J, Nadel H, Nieder M, Sato J, Siegal G, Weiner M, Wells R, Wold L, Womer R, Grier H. Osteosarcoma: a randomized, prospective trial of the addition of ifosfamide and/or muramyl tripeptide to cisplatin, doxorubicin, and high-dose methotrexate. J Clin Oncol. 2005;23(9):2004–2011. doi: 10.1200/JCO.2005.06.031. [DOI] [PubMed] [Google Scholar]
- Mirabello L, Troisi RJ, Savage SA. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int J Cancer. 2009;125:229–234. doi: 10.1002/ijc.24320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the surveillance, epidemiology, and End results program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari C, Medri L, Follo MY, Piazzi M, Mariani GA, Calistri D, Cocco L. PI-PLCβ1 gene copy number alterations in breast cancer. Oncol Rep. 2012;27(2):403–408. doi: 10.3892/or.2011.1529. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Takeuchi K, Muraoka S, Takezoe H, Takahashi N, Mori N. A neurally enriched coronin-like protein, ClipinC, is a novel candidate for an actin cytoskeleton-cortical membrane-linking protein. J Biol Chem. 1999;274(19):13322–13327. doi: 10.1074/jbc.274.19.13322. [DOI] [PubMed] [Google Scholar]
- Niggli V, Rossy J. Ezrin/radixin/moesin: versatile controllers of signaling molecules and of the cortical cytoskeleton. Int. J Biochem Cell Bio. 2008;40:344–349. doi: 10.1016/j.biocel.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Noh DY, Shin SH, Rhee SG. Phosphoinositide-specific phos- pholipase C and mitogenic signalling. Biochim Biophys Acta. 1995;1242:99–114. doi: 10.1016/0304-419x(95)00006-0. [DOI] [PubMed] [Google Scholar]
- O'Carroll SJ, Mitchell MD, Faenza I, Cocco L, Gilmour RS. Nuclear PLCbeta1 is required for 3T3-L1 adipocyte differentiation and regulates expression of the cyclin D3-cdk4 complex. Cell Signal. 2009;21(6):926–935. doi: 10.1016/j.cellsig.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Ochocka AM, Pawelczyk T. Isozymes delta of phosphoinositide-specific phospholipase C and their role in signal transduction in the cell. Acta Biochim Pol. 2003;50(4):1097–1110. [PubMed] [Google Scholar]
- Oka M., Hironori Edamatsu, Makoto Kunisada, Lizhi Hu, Nobuyuki Takenaka, Siphora Dien, Masanobu Sakaguchi, Riko Kitazawa, Kazumi Norose, Tohru Kataoka and Chikako NishigoriEnhancement of ultraviolet B-induced skin tumor development in phospholipase Cε-knockout mice is associated with decreased cell death. Carcinogenesis. 2010 Oct;31(10):1897–902 [DOI] [PubMed]
- Ou L, Guo Y, Luo C, Wu X, Zhao Y, et al. RNA interference suppressing PLCE1 gene expression decreases invasive power of human bladder cancer T24 cell line. Cancer Genet Cytogenet. 2010;200:110–119. doi: 10.1016/j.cancergencyto.2010.01.021. [DOI] [PubMed] [Google Scholar]
- Pujuguet P, Del Maestro L, Gautreau A, et al. Ezrin regulates E-cadherin-dependent adherens junction assembly through Rac1 activation. Mol Biol Cell. 2003;14:2181–2191. doi: 10.1091/mbc.E02-07-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SG. Regulation of phosphoinositide-specific phospholipase C. Annu Rev Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi P, Solimando L, Zini N, Santi S, Riccio M, Guidotti L. Inositol-specific phospholipase C in low and fast proliferating hepatoma cell lines. Int J Oncol. 2003;22:1147–1153. doi: 10.3892/ijo.22.5.1147. [DOI] [PubMed] [Google Scholar]
- Suh PG, Park J, Manzoli L, Cocco L, Peak JC, Katan M, Fukami K, Kataoka T, Yun S, Ryu SH. Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Rep. 2008;41:415–434. doi: 10.5483/BMBRep.2008.41.6.415. [DOI] [PubMed] [Google Scholar]
- Tan XG, Yang ZL. Expression of Ezrin, HGF, C-met in pancreatic cancer and non-cancerous pancreatic tissues of rats. Hepatobiliary Pancreat Dis Int. 2010;9(6):639–644. [PubMed] [Google Scholar]
- Tsukita S, Yonemura S. ERM (Ezrin/radixin/moesin) family: from cytoskeleton to signal transduction. Curr Opin Cell Biol. 1997;9:70–75. doi: 10.1016/S0955-0674(97)80154-8. [DOI] [PubMed] [Google Scholar]
- Yang L, Guo T, Jiang S, Yang Z. Expression of Ezrin, HGF and c-met and its clinicopathological significance in the benign and malignant lesions of the gallbladder. Hepatogastroenterology. 2012;59(118):1769–1775. doi: 10.5754/hge11744. [DOI] [PubMed] [Google Scholar]
- Zhao H, Shiue H, Palkon S, et al. Ezrin regulates NHE3 translocation and activation after Na+−glucose cotransport. Proc Natl Acad Sci U S A. 2004;101:9485–9490. doi: 10.1073/pnas.0308400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhang X, Xin Y. Up-regulated expression of Ezrin and c-Met proteins are related to the metastasis and prognosis of gastric carcinomas. Histol Histopathol. 2011;26(9):1111–1120. doi: 10.14670/HH-26.1111. [DOI] [PubMed] [Google Scholar]
- Zhu L, Zhou R, Mettler S, et al. High turnover of Ezrin T567 phosphorylation: conformation, activity, and cellular function. Am J Physiol Cell hysiol. 2007;293:C874–C884. doi: 10.1152/ajpcell.00111.2007. [DOI] [PubMed] [Google Scholar]