Table 3. KEGG pathway mapping.
| Module cluster → | 8 | 1 | 1 | 11 | 20 | 20 | 9 | 9 | Injury indicator |
| KEGG pathway ↓ Module → | 23 | 3 | 1 | 27 | 40 | 41 | 24 | 25 | |
| Biosynthesis of unsaturated fatty acids | ↑ | Albumin increase & Eosinophilia | |||||||
| Butanoate metabolism | ↑ | ||||||||
| Fatty acid metabolism | ↑ | ||||||||
| Peroxisome | ↑ | ||||||||
| Propanoate metabolism | ↑ | ||||||||
| Valine, leucine and isoleucine degradation | ↑ | ||||||||
| Drug metabolism - CYP P450 | ↓ | Spleen weight decrease | |||||||
| Metabolism of xenobiotics by CYP P450 | ↓ | ||||||||
| Glutathione metabolism | ↓ | ||||||||
| Glutathione metabolism | ↑ | Periportal lipid accumulation | |||||||
| Proteasome | ↑ | ||||||||
| Natural killer cell mediated cytotoxicity | ↑ | ↑ | Periportal fibrosis | ||||||
| Regulation of actin cytoskeleton | ↑ | ↑ | |||||||
| Leukocyte transendothelial migration | ↑ | ↑ | |||||||
| Phagosome | ↑ | ↑ | |||||||
| Pyruvate metabolism | ↓ | Alkaline phosphatase & Liver weight decrease | |||||||
| Pyruvate metabolism | ↓ | ↓ | Basophil increase |
Pathways are at left, injury indicators are at right, and connecting gene co-expression modules are in the middle. Pathways are included only if they are associated with absolute module activation scores, 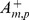 , greater than 1.5, if they have Bonferroni-corrected p-values less than 0.05, and if there are at least six hits from the module in the pathway.
, greater than 1.5, if they have Bonferroni-corrected p-values less than 0.05, and if there are at least six hits from the module in the pathway.
