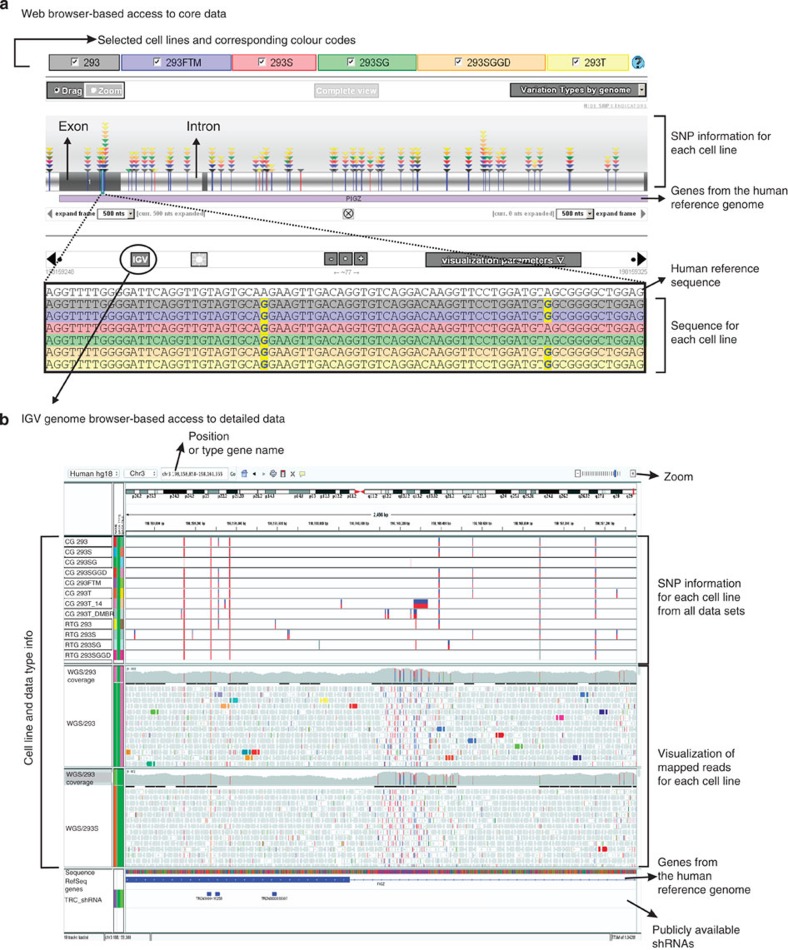
Figure 6. Visualization of SNPs and indels in the 293 Variant Viewer.
(a) Snapshot of the 293 Variant Viewer for the PIGZ gene. The upper region gives an overview of the gene with its variations in each genome, colour-coded by variation type and cell line. Triangles indicate the presence of the variant in a particular genome. The lower part of the browser allows detailed inspection of the sequence and comparison with the human reference genome. A link to the same region in IGV is provided as well. (b) Overview of SNP calling and realignment data tracks in the IGV genome browser for the same gene as in a. The two SNP calling algorithm tracks (CG and RTG) are shown with homozygous SNPs (red bar) and heterozygous SNPs (red/blue). In the CG tracks, no-calls are also shown in light red. In regions where the realignment coverage is zero, the sequence is the same as the human reference sequence. The TRC shRNA track allows the detection of SNPs in target regions of the shRNAs from the TRC2 collection (Broad Institute and Sigma). Mousing-over the different tracks provides users with extra information about specific features, such as mapping quality, base type count and phred scores.