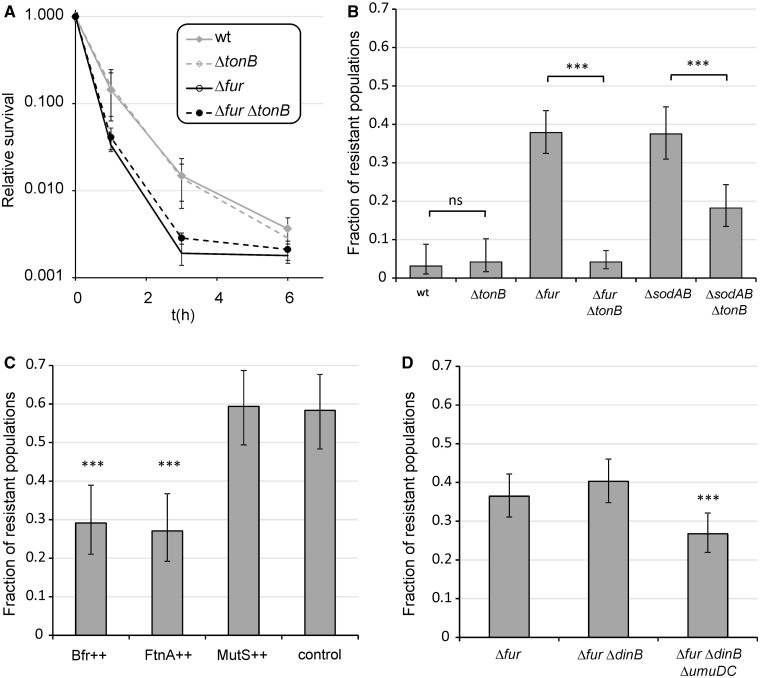
Fig. 3.
Impact of iron uptake, iron storage, and inactivation of error prone polymerases on ciprofloxacin-resistant evolution. (A) Effect of tonB deletion on survival under ciprofloxacin exposure in WT and Δfur genetic backgrounds. The mean of values is shown for n = 8. (B) Effect of impaired ferric iron uptake due to deletion of tonB gene on the fractions of ciprofloxacin-resistant populations in the case of Δfur and ΔsodAB. Fractions are based on a total of n = 96 (wt vs. ctonB), n = 288 (Δfur vs. ΔfurΔtonB), and n = 192 (ΔsodAB vs. ΔsodABctonB) populations of each genotype. Error bars represent confidence intervals of proportions (Wilson procedure). (C) Effect of overexpressing mutS and genes involved in iron storage (bfr, ftnA) on the fractions of ciprofloxacin-resistant populations in the case of Δfur. Fractions are based on a total of n = 96 populations of each genotype. Error bars represent confidence intervals of proportions (Wilson procedure). Genes of interests were overexpressed from a high-copy number plasmid (pZE31) in a Δfur genetic background. Control, Δfur harboring empty pZE31 plasmid; ++, overexpression of a given gene from the pZE31 plasmid. (D) Effect of the inactivation of error prone DNA polymerases PolIV (dinB) and PolV (umuDC) on the fractions of ciprofloxacin-resistant populations in the case of Δfur. Fractions are based on a total of n = 288 populations of each genotypes. Error bars represent confidence intervals of proportions (Wilson procedure). ***P ≤ 0.001; ns, P > 0.05 (chi-square test using the actual number of observations).