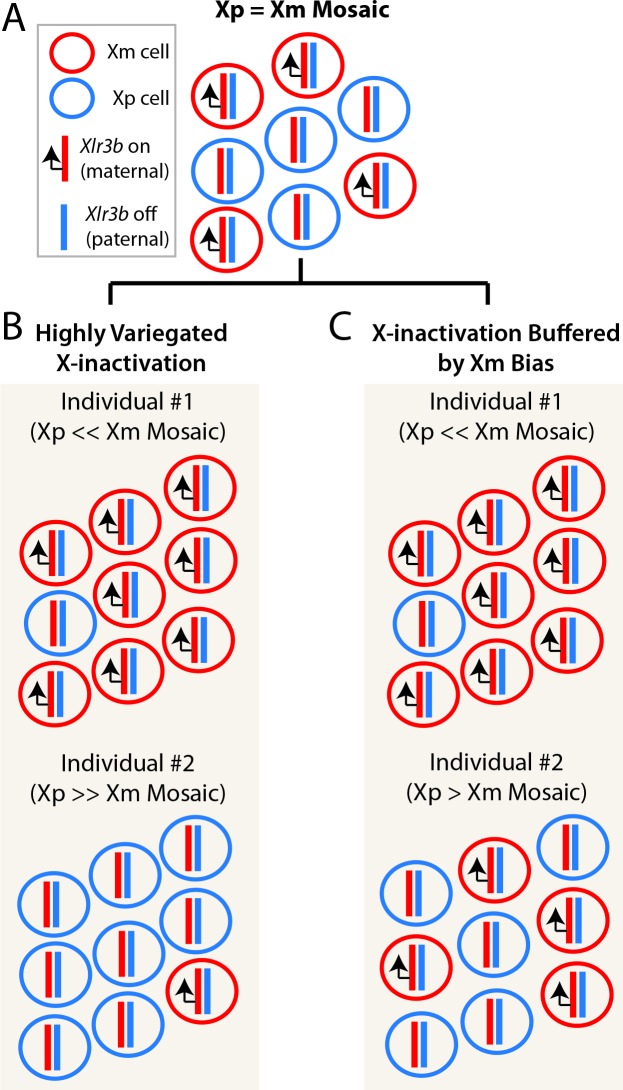
Figure 2. The relationship between X-linked imprinted genes and random X-inactivation patterns.
This schematic illustrates the relationship between different X-inactivation patterns and X-linked imprinted genes, such as Xlr3b, which is an X-linked imprinted gene that is preferentially expressed from the Xm allele. (A) If random X-inactivation in female tissues resulted in an equal number of Xm and Xp expressing cells, then X-linked imprinted genes like Xlr3b would have a consistent effect on different individuals. However, this pattern does not occur. (B) The study by Wu and colleagues [7] uncovered highly variable X-inactivation patterns in different tissues and individuals. This random and variegated pattern results in some brain regions or tissues being primarily populated by Xm+ cells in one individual and Xp+ cells in another. This pattern would introduce extreme variance for the effects of X-linked imprinted genes, like Xlr3b, in a population of individuals. (C) A parent-of-origin effect that biases X-inactivation toward the Xm could potentially buffer the variance associated with random X-inactivation, such that a minimum contribution of Xm+ cells exists in tissues where Xlr3b imprinting functions, such as the brain. In this scenario, pronounced Xm biases would still arise, perhaps increasing the impact of Xlr3b imprinting effects (individual #1), but the Xm inactivation bias would buffer against extreme Xp biases that could negate the effects of Xlr3b imprinting (individual #2).