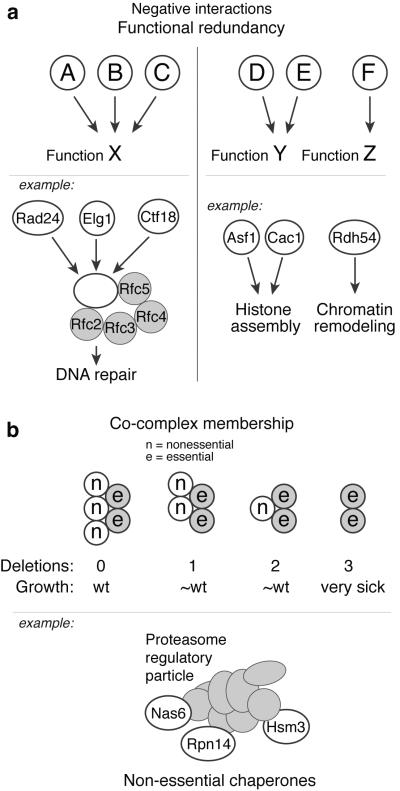
Figure 2. Negative genetic interactions revealed by TMA.
A) Functional redundancy: (left panel) Three factors (A, B and C) carry out function X in a redundant fashion, but pair-wise deletions (e.g. AΔ × BΔ) in an E-MAP will not reveal this as the exacerbation is covered by the remaining wild-type factor. Deletion of all three factors (e.g. AΔ BΔ × CΔ) will however reveal the exacerbating relationship. For example, three replication factor C (RFC)-like protein complexes play key roles in maintaining genome stability. These complexes share four subunits (Rfc2, Rfc3, Rfc4 and Rfc5) but differ in their large subunit (Rad24, Elg1 or Ctf18), and function in parallel DNA repair pathways. (right panel) Two factors (D and E) carry out the same function Y, and deletion of either alone will not disrupt Y. Hence, standard E-MAPs (e.g. DΔ × FΔ) will miss connections between Y and other functions (Z). Conversely, TMA (DΔ EΔ × FΔ) will properly disrupt Y and be able to detect genetic interactions with other functions. The Swi/Snf Rdh54 protein was found to compensate for loss of the histone chaperones Asf1 and Cac1 using TMA. B) Co-complex membership: Non-essential components of the same protein complex can act in a redundant fashion so that deleting a subset of these has little effect. In the depicted schematic, standard E-MAPs would not detect co-complex members, as a single or double deletion results in near-wild-type phenotype. TMA would however allow for deleting two known complex members and cross these against candidate deletions, uncovering a synthetic sick interaction with the third deletion. The three proteasome regulatory particle chaperones Nas6, Rpn14 and Hsm3 have little effect on fitness at 30°C when deleted individually, whereas the triple deletion adversely affects cell growth.