Figure 1.
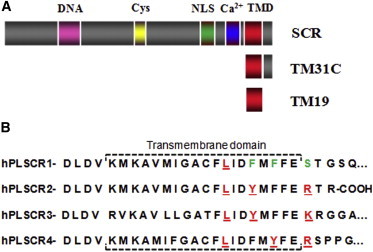
(A) SCR domains and sequence-derived peptides: DNA-binding domain (pink), Cys-rich domain (yellow), nuclear localization signal (green), calcium-binding domain (blue), and TMD (red). (B) Representation of the human phospholipid (hPL) SCR family putative TMD sequence. hPL SCRs 2, 3, and 4 contain a CRAC motif ([L/V]-[X](1-5)-[Y]-[X](1-5)-[R/K]) localized at the C-terminal end of the predicted transmembrane helix (amino acids in red), whereas SCR presumably is the result of several mutations (in green), such as the polar amino acid (serine) instead of a positively charged amino acid (arginine or lysine), and two phenylalanines in different positions, which appear as tyrosines in the rest of the family sequences. Thus, neither the CRAC nor the CRAC-like sequence ([L/V]-[X](1-5)-[F]-[X](1-5)-[R/K]) is completed in SCR TMD. Sequence information was taken from UNIPROT. Sequences were aligned according to Sahu et al. (3) and Bateman et al. (7).
