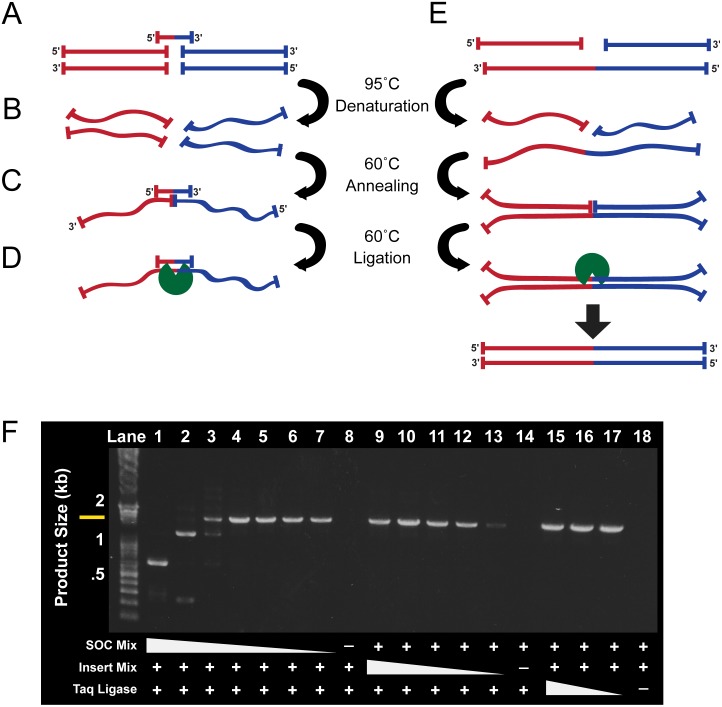
Figure 1. Cycled Ligation Assembly Reaction.
(A) Diagram of the assembly of two double-stranded DNA fragments (red and blue) using a Scaffold Oligonucleotide Connector (SOC). (B) DNA fragments are denatured at 95°C in a thermocycler instrument. (C) The reaction is cycled to 60°C, allowing a SOC homologous to the last 20 bp of the first DNA fragment and the first 20 bp of the second fragment to anneal, forming a transient partly double-stranded (ds) DNA structure. (D) A thermostable ligase (green) specific for nicks in dsDNA seamlessly ligates the two bottom strands. (E) Subsequent cycles of denaturation and annealing/ligation allow the thermostable ligase to ligate the top strands, using the ligated bottom strands as a template. Further cycles of ligation exponentially increase the amount of correct product as the products of early rounds served as an expanding pool of template for later cycles. (F) Successful assembly of three ∼0.6 kb DNA inserts ( Table 1 ) using a cycled ligation assembly was confirmed by PCR amplification of a correctly sized 1.8 kb fragment (yellow bar). A range of concentrations of reaction components was tested in order to find optimal conditions. SOC concentration (ten-fold serial dilutions from 10 µM to 10 pM, lanes 1–7), inserts concentration (6 nM, 2 nM, 0.6 nM, 0.2 nM, 0.06 nM, lanes 9–13), and Taq Ligase concentration (240 U, 80 U, 24 U, lanes 15–17) were varied (+ denotes optimal concentrations of 10 nM SOCs, 2 nM inserts, and 80 U taq Ligase).