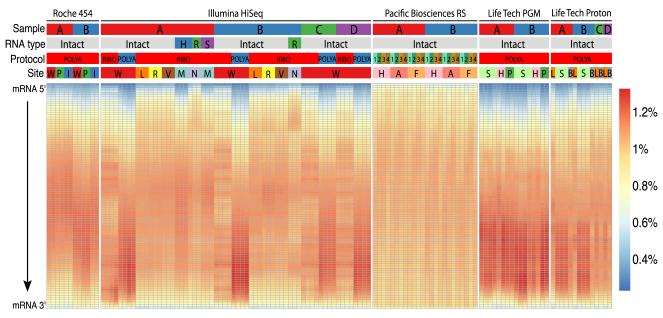
Figure 2. Transcript coverage across all genes detected.
Each gene was examined as a set of 100 adjacent segments (percentiles of total transcript length). The relative number of reads that map to each segment was then plotted for each sample, platform, and technique (percent of all library reads per segment, see heatmap color key). Samples are categorized by five parameters (top): NGS platforms: Roche 454 GS FLX+, Illumina HiSeq 2000/2500, Pacific Biosciences RS, Life Technologies PGM and Proton; input RNA sample: samples A (red), B (blue), C (green), and D (purple); RNA type: intact or degraded by heat (H, blue), RNase (R, green) or sonication (S, purple); library protocol: polyA enrichment, ribosomal RNA depletion (ribo) or polyA plus 5′ cap enrichment with (1, 2, 3) or without (4) size fractionation; and site: 14 core facility sequencing laboratories. Most platforms showed less coverage at the 5′ and 3′ ends of the transcripts. Details on sequencing platforms, site abbreviations, sample type chemistries, and library preparations are listed in Table 2.