Abstract
The development of a hematopoietic reporter is crucial for determining the fate of lineages derived from cell-based therapies. A marking system will enable safer embryonic stem (ES) and induced pluripotent stem (iPS) cell-based derivation of blood lineages, and facilitate the development of efficient cellular reprogramming strategies based on direct fibroblast conversion. Here we report that the protein tyrosine phosphatase CD45 is an ideal candidate gene on which to base a hematopoietic reporter. CD45 regulatory elements were discovered by analyzing transcription factor chromatin occupancy (ChIP-seq) and promoter nuclease sensitivity (DNase-seq) to identify minimally sufficient sequences required for expression. After cloning the CD45 regulatory elements into an attenuated lentiviral backbone, we found that two transcriptional initiation regions were essential for high-level expression. Expressing CD45 promoters containing these regions and tethered to GFP in a primary B cell differentiation assay and a transplantation model resulted in high levels of GFP in lymphoid, myeloid and nucleated erythroid cells in mouse and human blood cell lineages. Moreover, high GFP levels remained five months following secondary transplantation, indicating persistence of the reporter. No CD45 driven GFP expression is observed following fibroblast or ES cell transduction. The GFP reporter is seen only after ES cells differentiate into hematopoietic cell progenitors and lineages, suggesting that this hematopoietic reporter system could be useful in validating potential autologous blood cell therapies.
Introduction
An effective treatment regimen for hematologic disease and malignancy has been challenging due to a lack of suitably-matched donors (1). To circumvent this issue, efforts have focused on generating hematopoietic stem and progenitor cells (HSPCs) from embryonic stem (ES) (2, 3) and induced pluripotent (iPS) cells (4, 5). However, the wide-spread application of pluripotent stem cells is currently hampered by their tumorigenic potential. A proposed alternative is the direct conversion of fibroblasts into HSPCs and blood cells (6, 7). Although lineage specification and reconstitution potential are currently inefficient (6, 7), evidence suggests that improvements in direct hematopoietic reprogramming could provide a viable strategy for hematopoietic based therapeutics. Aided by GFP reporters, recent studies demonstrated that overexpression of specific transcription factors facilitated generation of neurons and cardiomyocytes from fibroblasts (8, 9), suggesting the conversion of fibroblasts into functional HSPCs was plausible. These studies highlight the importance of having a reporter system for hematopoietic marking and a method to track cellular reprogramming.
Since the blood cell therapy field lacks a reliable reporter for hematopoietic production after differentiation of ES and iPS cells (5, 10), the development of a hematopoietic restricted marking system is essential. Furthermore, a fluorescent reporter system enables real time tracking of full reprogramming (11, 12), permits the study of reprogramming intermediates (13, 14), and may facilitate the eventual use of small molecules for direct reprogramming, as demonstrated recently for iPS cell derivation (15). Additionally, a reporter construct could aid in the purification and removal of undifferentiated pluripotent cells to minimize teratoma formation upon transplantation.
An effective reporter should be inactive in fibroblasts and pluripotent stem cells, but turned on in the desired reprogrammed cell fate. Transcription factors such as Gata2, Hoxb4 and Evi1 were previously employed as reporters due to their essential roles in HSPC genesis, maintenance and/or amplification (16-20). However these reporters were not limited to blood cell lineages, and this limited their utility (21, 22). Additionally, these transgenes used in the production of reporter mice (23) cannot be virally introduced into hematopoietic reconstituting cells because the reporter is too large for the viral backbone (24).
In this study we chose the transmembrane protein CD45 as the foundation for our reporter. CD45 (also known as Ly-5, B220 and Ptprc) is highly abundant on the cell surface of all nucleated blood cells, but absent on other cell types (22, 25). This receptor is expressed early during hematopoietic development (26). CD45 expression increases as HSPCs differentiate and transcript expression depends on the developmental stage, lineage specified, and activation state of the cell. Previous work has identified two or more promoters that initiate transcription from one of two alternate starting exons or the first intron, with all transcripts sharing a translational start on exon 2 (26). Prior studies employing retroviral delivery were unable to document CD45 promoter activity using 0.8kb and 2.8kb promoter fragments (27). However, Virts et al. showed modest expression of a CD45 minigene driven by an 839bp DNA element in stably transfected cytotoxic T cells following drug selection (28). Additionally, a CD45-YFP reporter mouse was created by Yang et al. that facilitated visualization of hematopoietic cells in vivo (29). This mouse model enabled genetic tracking studies; but since it was a knockin at the translational initiator codon, the identification of minimally sufficient reporting elements required for CD45 marking was precluded. Critically, a virally transducible reporting system that can mark multilineage human hematopoiesis has not yet been published. Based on regulatory elements identified using ChIP-seq and DNase-seq data, we demonstrate that CD45 expression is controlled by a small region that is crucial for driving maximal expression of a GFP transgene. Here we report that CD45 can serve as a highly specific reporter to efficiently mark multilineage hematopoiesis.
Methods
Cell lines
Mouse hematopoietic progenitor cell (EML), B cell (CH12.LX), nucleated erythroid progenitor (MEL); and human HSPC (KG1a), B cell (Raji), T cell (Jurkat) and nucleated erythroid (K562) cell lines were used for initial CD45 reporter testing studies.
Mouse strains
C57BL/6 mice with the CD45.2/ly5.2/Ptprcb haplotype and congenic C57BL/6.SJL mice with the CD45.1/ly5.1/Ptprca haplotype were obtained from Jackson laboratory. All mouse experiments were in compliance with protocols approved by the Institutional Animal Use and Care Committee of the University of Iowa.
RNA isolation and quantitative real-time PCR (qPCR)
Bone marrow was harvested and lysed with 1xBD Pharm Lyse and fresh RNA was isolated using the Illustra RNAspin Mini RNA Isolation Kit. cDNAs were synthesized using Promega GoScript™ Reverse Transcriptase according to manufacturer recommendations. The cDNA products were then diluted 40-fold, and 5μl was used for each qPCR reaction following amplification with Takara Ex Taq polymerase HS. Primers for CD45, CD34, Vav1, Myb, Gapdh, and Hprt1 were employed (see Table 1 in Supplemental Methods for details). Detection was performed with a Biorad CFX96 real-time PCR machine.
Table 1. List of primers used in this study.
| Reporter construction primers (AttB sequences in red, genomic hybridizing sequences in blue) | |
|---|---|
| CD45 prom −164 AttB1-F | ggggacaagtttgtacaaaaaagcaggcttacctttagaggaaaattgagacga |
| CD45 prom +168 AttB2-R | ggggaccactttgtacaagaaagctgggtaaataatggttatctctcttgaagtttg |
| Quantitative real-time PCR primers | |
| CD34 qRT-PCR-F | ggtagctctctgcctgatgagt |
| CD34 qRT-PCR-R | ttggtaggaactgatggggata |
| CD45 qRT-PCR-F | ggcagatgatattccaaagaaa |
| CD45 qRT-PCR-R | atctccacttccatgtctccat |
| Myb qRT-PCR-F | gagatgtgtgaccatgactacga |
| Myb qRT-PCR-R | gttctgttccaccagcttcttc |
| Vav1 qRT-PCR-F | gcagtgaactctttgaggcttt |
| Vav1 qRT-PCR-R | gattcctttgttctgggcaat |
| Hprt1 qRT-PCR-F | aactttgctttccctggttaa |
| Hprt1 qRT-PCR-R | tcaagggcatatccaacaaca |
| Gapdh qRT-PCR-F | tgtgtccgtcgtggatctga |
| Gapdh qRT-PCR-R | cctgcttcaccaccttcttga |
| Lentiviral vector titering primers | |
| LV WPRE-F | accacctgtcagctcctttc |
| LV WPRE-R | caacaccacggaattgtcagt |
| LV VSV-G-F | ccattattgcccgtcaagctc |
| LV VSV-G-R | ggagtgaaggatcggatggact |
| LV gag-F | atgggtgcgagagcgtcagt |
| LV gag-R | caggccaggattaactgcga |
Gene expression and regulatory element identification
Please see Supplemental Methods for details.
Reporter construction
The eight truncations of the CD45 promoter region positioned from −164 to +168 of the transcriptional start site were cloned into the lentiviral vector driving EGFP (please see supplemental methods for details). For HM1 ES cell transduction, EGFP was replaced with mCherry to enable co-transduction with a HOXB4-GFP expression cassette.
Lentivirus production
The lentiviral vector used in these studies has been described previously (24), and contains a self-inactivated (SIN) long terminal repeat (LTR), Woodchuck hepatitis virus post-transcriptional Regulatory Element (WPRE) and central polypurine tract (cPPT) to increase biosafety, full-length RNA production and titer, respectively. Please see Supplemental Methods for further details.
In vitro B cell differentiation assay
Please see Supplemental Methods for details.
ES cell differentiation
HM1 ES cells harboring a HOXB4-GFP expression cassette (30) were transduced with a CD45 reporter driving mCherry for 24hr prior to differentiation as previously described (4, 31). Briefly, ES cells were subjected to hanging drop formation in culture for 2 days to form embryoid bodies (EB), followed by 4 days in suspension. On day 6, the EBs were dissociated and co-cultured with the OP9 stromal cell line and supplemented with the hematopoietic cytokines mSCF, mVEGF, hTPO and hFLT3L. At day 10, cells were stained with antibodies against c-kit, Sca-1, CD45, and Gr-1/Mac-1.
Transplantation
Bone marrow from CD45.1 mice was harvested and lineage depleted with biotin conjugated monoclonal antibodies (B220, CD4, Gr-1, Mac-1, NK1.1, CD8). Subsequently, cells were cultured overnight in D-MEM supplemented with 10% FBS, mSCF, hTPO, and hFLT3L. After 24hr, 1×106 cells were transduced with a multiplicity of infection (MOI) of 30 in the presence of 8μg/ml polybrene and incubated overnight. Next, 2×105 cells were transplanted into congenic CD45.2 mice that had been irradiated with a 950 cGy dose. In transplants receiving supportive bone marrow, 1×104 untransduced cells from the donor were also supplemented. Secondary transplants received 1×106 bone marrow cells from primary donor recipients.
Results
Selection of a hematopoietic reporter gene
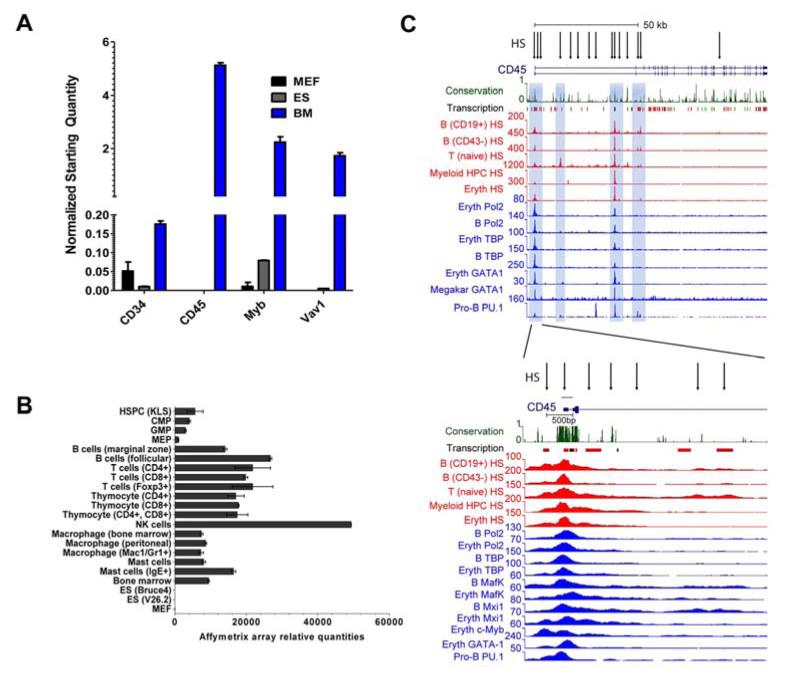
To find a suitable hematopoietic reporter gene, we examined gene candidates that were highly expressed in bone marrow cells, but excluded in other cell types. By mining publically available expression profiling data (22, 32), we evaluated the levels of well-established hematopoietic markers, including CD45, CD34, Myb, and Vav1. The expression of these candidates was verified by qRT-PCR, as shown in Figure 1A. CD45 levels were as anticipated significantly higher than any of the other genes, since this protein makes up approximately ten percent of protein on the surface of hematopoietic cells (33). Importantly, CD45 levels were high in a panel of mouse and human blood cell lineages that included representatives of the lymphoid, myeloid and nucleated erythroid compartments, as well as the multipotent progenitors and HSPCs that specify these cell types (Figure 1B and Supp Figure 1). Levels of the hemato-endothelial marker Vav1 were approximately 10-fold lower than CD45 in all mouse and human hematopoietic lineages (Figure 1A). CD34 is a marker widely used for human HSPC isolation (34); however, our initial study using reporters based on CD34 and Sca-1 promoter showed that their promoters were not as strong and as tissue specific when compared to CD45 (results not shown). The data suggested that CD45 was a strong hematopoietic reporter candidate.
Figure 1. Hematopoietic reporter candidates, CD45 regulatory element discovery and promoter characterization.
(A) Quantitative real-time PCR (qRT-PCR) expression levels of highly hematopoietic or hemato-endothelial (Vav1) enriched genes. Murine embryonic fibroblasts (MEF), embryonic stem (ES) cells and whole bone marrow were analyzed. Levels were normalized to Hprt and Gapdh. (B) Affymetrix array measured CD45 expression levels in hematopoietic progenitor and differentiated cell lineages. Kit+, lineage-, Sca-1+ (KLS), Common myeloid progenitor (CMP), Granulocyte-macrophage progenitor (GMP), and Megakaryocyte-erythroid progenitors (MEP) are also indicated. Embryonic stem (ES) and murine embryonic fibroblasts (MEF) are included as non-hematopoietic lineages that do not express CD45. (C) The full-length CD45 gene with its two most common alternative transcripts is shown in the top panel, and an enlarged view of the proximal promoter region can be seen at the bottom. Exons 1b and 2 are drawn as blue boxes. The region mapped for promoter activity spanning −164 to +168 (−164/+168) is shown with a green line over it. Only the 15 HS that were shared among 3 or more of the 6 cell lines with DNase-seq data are shown. Of these 15 HS, 6 were shared across all 6 lines, and all but 1 were shared by 4 or more cell linesVertebrate conservation and Affymetrix exon array enrichment for hematopoietic tissues (labeled Transcription) are indicated immediately below the gene structure, with lines in red depicting high-level expression. Hematopoietic cell regulatory elements marked by DNase hypersensitivity (HS) and transcription factor occupancy were obtained from ChIP-seq data. HS are marked by arrows.
CD45 gene regulatory element identification
Next, we analyzed global transcription factor occupancy following ChIP-seq and open chromatin from DNase-seq to discover the regulatory elements that controlled CD45 expression in the mouse (Figure 1C and Supp Figure 2) and human CD45 gene (Supp Figure 3). DNase hypersensitivity sites (HS), transcription factor and RNA polymerase II chromatin occupancy, and the histone H3 lysine 4 trimethylation (H3K4me3) mark for gene activation were determined in mature and progenitor B cell subsets, unstimulated T cells, nucleated erythroid cells (MEL), and a CD34 positive multipotent myeloid progenitor cell (416B) line (Figure 1C).
We focused our attention on the strong HS observed at the proximal promoter region encompassing the downstream alternate exon 1 as this is where expression of most CD45 transcription initiates from. We identified HS and chromatin occupancy of essential hematopoietic transcription factors that mark the CD45 core promoter region and overlap exon 1 and intron 1, (Figure 1C). PU.1, GATA1, c-Myb, Mafk and Mxi1 are all well-known regulators of multilineage blood cell specification and their high level enrichment can be observed at this region. GATA2, Tal1, and other regulators of HSPC specification were also found enriched within the CD45 gene and proximal promoter region (Supp Figure 2). Significant Pol2 and TATA-binding protein (TBP) occupancy was observed as expected and helped define the critical promoter for analysis. Collectively, the DNase-seq and ChIP-seq data confirmed that the region of interest most crucial for our CD45 reporter is largely confined to exon 1 and the first intron.
CD45 promoters efficiently mark mouse hematopoietic cells
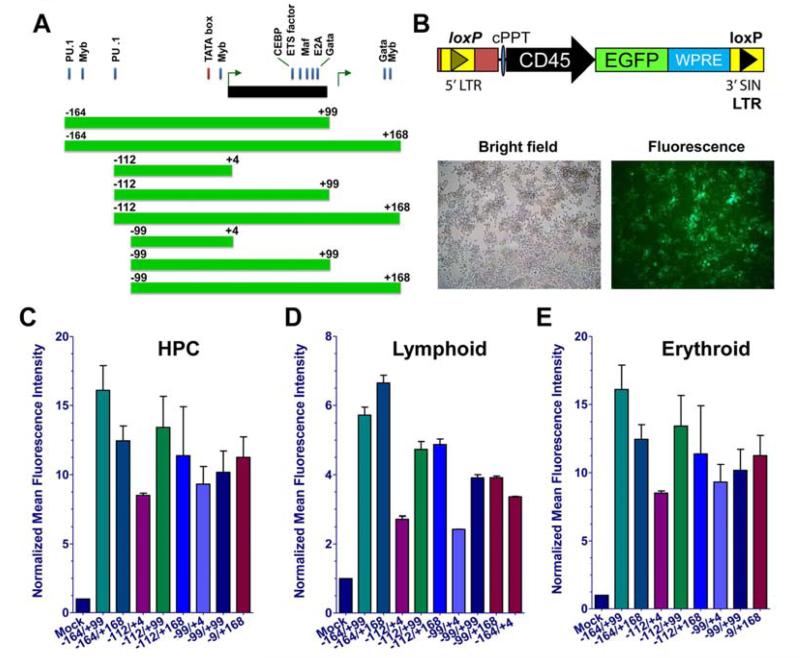
We constructed lentiviral GFP reporters with 8 different CD45 promoter truncations to determine the minimal regulatory element configuration required to mark hematopoietic cells (Figure 2A). The regulatory region spanning promoters P1b, intron 1 and P2 (−164 to +168; −164/+168) contained dual PU.1, c-Myb and GATA binding sites, as well as other critical regulatory footprints. We used our same lentiviral transduction system employed previously for long-term (5-7 months) in vivo GFP marking and delivery of therapeutic protein (24, 35). To test CD45 promoter strength we used a hematopoietic progenitor cell (EML), B cell (CH12.LX), and an erythroid progenitor (MEL) cell line. EML and CH12.LX lines are notoriously difficult to transduce, but we were able to demonstrate high level marking in both lines that could be readily detected by fluorescence microscopy (Figure 2B). Following reporter introduction into the multipotent progenitor, B cell and erythroid cell lines, the eight CD45 reporters regions resulted in similar frequencies of GFP positivity (Supp Figure 4), but notable differences in GFP mean fluorescence intensity (MFI) levels emerged that enabled us to better assess promoter activity. Flow cytometric analysis demonstrated that constructs consisting of regions −164 to +99 (−164/+99), −164 to +168 (−164/+168) and −112 to +168 (−112/+168) had the highest GFP intensity (Figure 2C-E). Minimal constructs, including reporters with promoters comprising only −112 to +4 or −99 to +4, were less efficient in promoting high GFP levels. We also stained the mouse lymphoid (CH12LX) and HPC (EML) cells with a CD45 antibody to corroborate that these lines express CD45 at a high level. Although 95-99 percent of the cells expressed CD45, levels of marking only reached a level of 92 percent for the lymphoid line and a more modest level for HPCs (Supp Figure 5), likely reflecting incomplete transduction in these cell lines. Our findings corroborate the requirement for sequences that are bound by GATA1, c-Myb, Mafk, Mxi and PU.1 (36) to enable maximal promoter strength and GFP marking (see Figure 1C).
Figure 2. CD45 reporters mark mouse hematopoietic cell lines.
(A): Promoter design scheme is shown for CD45 reporter truncations based upon critical transcription factor binding sites. (B): Lentiviral vector design (top) used to transduce cells is shown. Representative images of a transduced B cell line following bright field and fluorescence microscopy (bottom) are depicted. The geometric mean was used to measure MFI, and GFP levels are normalized to mock as presented. (C): lymphoid B cell line (CH12Lx), (D): hematopoietic progenitor cell (HPC) line (EML), and (E): nucleated erythroid cell line (MEL). Transductions were done in duplicate and repeated with a minimum of 3 independent times. Abbreviations: MFI, mean fluorescence intensity; GFP, green fluorescent protein.
CD45 promoter activity is evolutionarily conserved in human hematopoietic cells
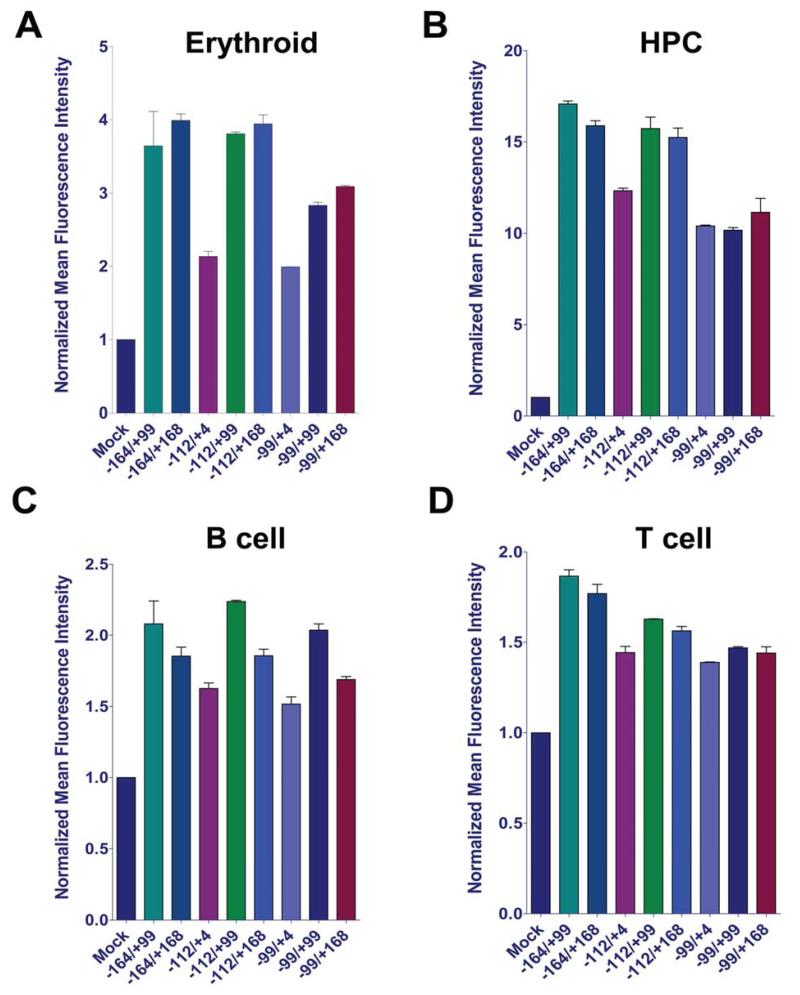
Since the CD45 promoter region of study is well-conserved, we tested the efficiency of reporter marking in human cells. The eight CD45 reporters were used to transduce human multipotent progenitor (KG-1), nucleated erythroid (K562), T cell (Jurkat) and B cell (Raji) lines (Figure 3, Supp Figures 6 and 7). Constructs lacking upstream and/or downstream regulatory elements were generally weaker across the different cell lines. Regions between −164 to +99, and −164 to +168 delivered maximal GFP levels, confirming the trend observed in mouse cell lines. The downstream PU.1 site and transcription factor binding sites resident in exon 1b were essential for maximal GFP expression. To ensure that our hematopoietic reporters would be suitable for human cellular reprogramming strategies, we replaced the mouse promoter sequence (−164 to +168) with the human promoter element following a multi-vertebrate species alignment (data not shown). We used the human CD45 reporter to transduce the human multipotent progenitor and B cell lines (Supp Figure 7, panels E-F). GFP levels observed were comparable to or exceeding those documented following transduction with the mouse reporters. We note that reporter strength may be underestimated due to lower CD45 promoter outputs from several of the human cell lines we used here, as documented by their lower CD45 expression levels (see Supp Fig 1). Collectively, marking measured by GFP intensity exhibited a similar trend to the mouse cell lines, indicating that the regulatory elements utilized are evolutionarily conserved.
Figure 3. CD45 reporters mark human hematopoietic cell lines.
(A): Normalized MFI calculated from flow cytometry for erythroid, (B): HPC, (C): B cell (C), and (D): T cell lines. Transductions were done in duplicate and repeated a minimum of 3 independent times. Abbreviations: MFI, mean fluorescence intensity; HPC, hematopoietic progenitor cell.
CD45 reporter is active in primary myeloid and lymphoid blood cell lineages
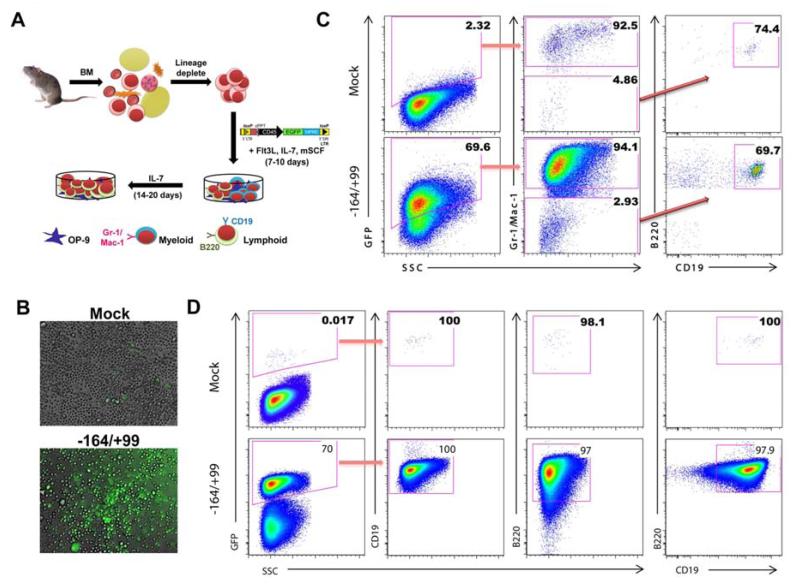
Based on our results from transduction of hematopoietic cell lines, we selected the - 164/+99 promoter driven CD45 reporter to assess reporting in a primary B cell culture derived from mouse BM as illustrated in Figure 4A. The enriched HSPCs from BM were transduced and stimulated with cytokines to differentiate them into myeloid and lymphoid lineages (37, 38). After 10 days, B cells (B220), pro-B cells (CD19), and myeloid cells (Gr-1/Ma-1) were assessed. High-level GFP marking and expression were observed with the CD45 reporters via confocal microscope (Figure 4B). The reporter marked approximately 70% of these cells as seen in the first panel of Figure 4C. This population was subgated to show 94% of the cells were Gr-1/Mac-1 positive (myeloid markers) in the middle panel. The remaining Gr-1/Mac-1 negative cells were B220 and CD19 positive. The reporter transduced cells exhibited the strong GFP levels that would be required for an effective reporter. To promote robust B cell differentiation, SCF was removed from the culture medium. At day 24 these cells were assayed again for B cell identity; as expected, complete B cell differentiation was observed. The ratio of GFP positivity remained unchanged with the exception that there was a much better separation between the non-transduced cells (negative), and transduced cells (positive). All of the GFP positive cells were mature B cells, as demonstrated by double positivity for B220 and CD19 (Figure 4D). The data suggest efficient reporting in primary B cells.
Figure 4. CD45 promoter enables primary myeloid and lymphoid cell reporting.
(A): Schematic of in vitro B cell differentiation assay. Mouse BM was isolated, lineage depleted, and stimulated with cytokines for the generation of myeloid cells (day 10) and B cells (day 20). (B): Mock and CD45 reporter fluorescent images were captured at 20X magnification with a confocal microscope. (C): Bone marrow derived B cell differentiation assay was used to document Gr-1/Mac-1 (myeloid), and B220/CD19 (lymphoid) surface staining 10 days following transduction. Cells were gated first for GFP positivity and subsequently this population was sub-gated (red arrows) for myeloid (Gr-1/Mac-1) and lymphoid (B220/CD19) lineages. (D): By day 24 of B cell differentiation, a uniform population of CD19 and B220 double positive B cells can be observed. Abbreviations: BM, bone marrow.
CD45 promoter activity is restricted to hematopoietic cells
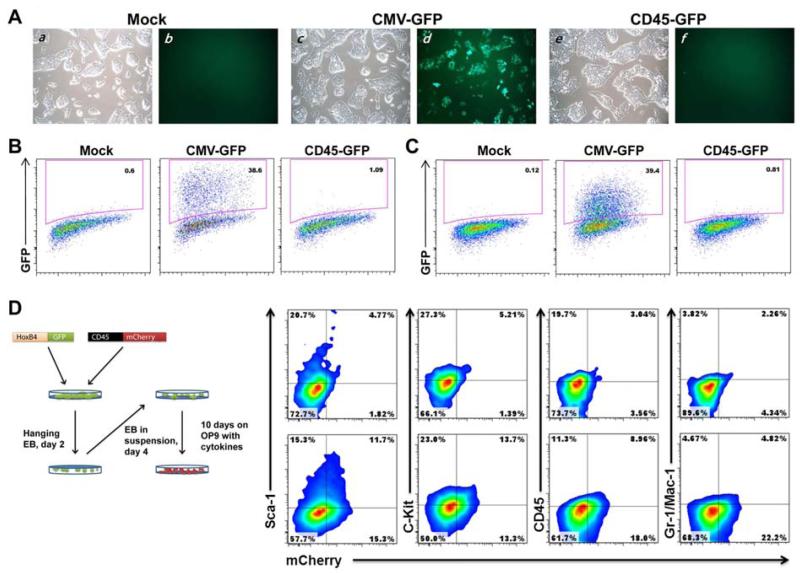
To validate the tissue specificity of our CD45 reporter activity, we transduced fibroblasts and ES cells; neither should be permissive for CD45 reporting potential based on gene expression analyses (see Figure 1). These cell types were selected because they are the substrate for direct reprogramming (6, 7) and pluripotent cell based differentiation (2, 3), respectively. After a 48 hour transduction period, cells were assayed for GFP expression using FACS. Transduction of mouse embryonic fibroblasts (MEF) with a ubiquitous CMV promoter at low multiplicities of infection (MOI 1) typically used in reprogramming experiments resulted in approximately 40% marking (Figure 5B). In contrast, our strongest CD45 reporter did not result in marking above baseline levels even 5 days post transduction (data not shown). Similarly, no evidence of GFP marking was observed by the CD45 reporter in ES cells (Figure 5C, compare CMV driven EGFP in the top right and CD45 in the bottom panels). To further demonstrate that CD45 promoter activity is restricted to blood cell lineages, ES cells that were transduced with the CD45 reporter were differentiated into HSPCs via embryoid body formation and subsequent OP9 stromal line co-cultivation (Figure 5D). Following EB formation and 10 days on OP9, cells were assayed for the presence of the cell surface hematopoietic markers Sca-1, c-Kit, CD45, and Gr-1/Mac-1. Sca-1 and c-Kit expression represents enrichment of HSPCs and Gr-1/Mac-1 serves as the predominant marker of the myeloid lineage. Prior to differentiation, these ES cells were transduced with HOXB4-GFP for increasing hematopoietic specification following differentiation. The HOXB4-GFP marker also facilitates isolation of ES cell derived lineages from the OP9 stromal layer. Since the GFP marker was embedded in the HOXB4 expression cassette, we employed an mCherry reporter to detect CD45 positivity in these experiments. HOXB4-GFP transduced cells were then subgated on mCherry and assessed for lineage marker expression. At this early time point approximately 27% of transduced cells were mCherry positive and these cells exhibited significant marking for Sca-1, c-kit, CD45, and Gr-1/Mac-1. These results highlight the multilineage marking capability of the CD45 reporter after ES cells are differentiated.
Figure 5. CD45-based hematopoietic reporter is not active in ES cells and fibroblasts.
(A): ES cells with mock (panels a, b), ubiquitous (CMV-GFP; panels c, d) or hematopoietic lineage restricted (CD45-GFP (−164/+99 construct); panels e, f) reporter transduction following brightfield (panels a, c, e) and fluorescent (panels b, d, f) imaging. (B): Flow cytometric measurement of MEFs and (C): ES cells following transduction with the designated lentiviral reporters. Note the lack of ES cell GFP fluorescence with the CD45 reporter. (D): HPC generation from ES cells. The strategy for ES cell differentiation is shown on the left. Sca-1, c-Kit, CD45 and Gr-1/Mac-1 surface staining (right) on HPCs and hematopoietic lineages following CD45 reporter (mCherry) and HOXB4-GFP transduction of ES cells. Mock (top) and −164/+99 CD45 reporters (bottom) are shown. These cells were analyzed at day 10 following co-culture on OP9 stromal cells. GFP positive cells were sub-gated for mCherry positivity. Abbreviations: ES, embryonic stem; MEF, murine embryonic fibroblast; HPC, hematopoietic progenitor cell; mCherry, monomeric cherry fluorescent protein.
CD45 reporters efficiently mark blood cells following transplantation
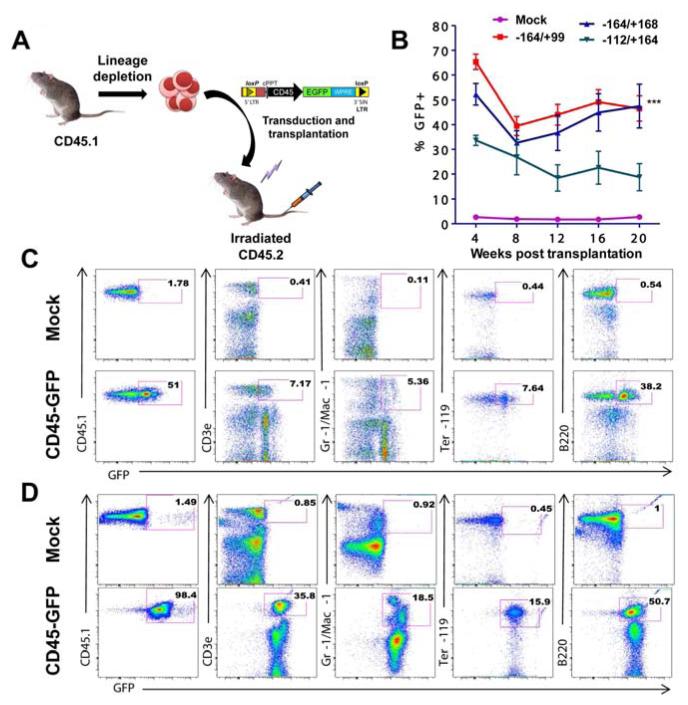
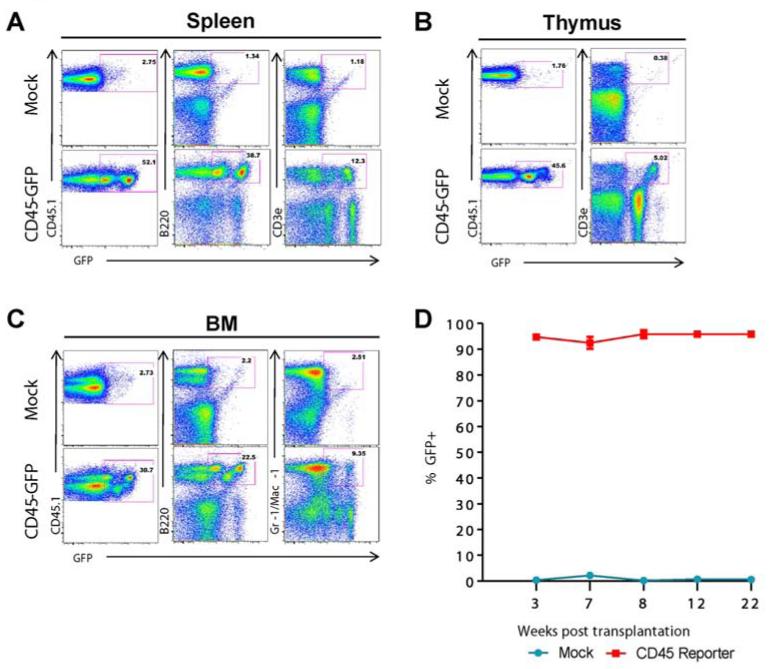
To confirm that our CD45 reporter could effectively mark multilineage hematopoiesis in vivo, bone marrow from CD45.1 mice (C57BL/6.SJL) was harvested and enriched for HSPCs. We transduced lineage depleted HSPCs with the three strongest CD45 reporters. As expected, approximately seven percent of our lineage negative population exhibited double positivity for Sca1 and c-Kit (Supp Figure 8), confirming enrichment of an HSPC population. HSPCs were injected into lethally irradiated CD45.2 mice, and reconstituted the host blood system by 4 weeks post transplantation (Figure 6). For our initial experiments we supplemented CD45 reporter transduced HSPCs with non-transduced supportive bone marrow to facilitate full transplant recovery. This resulted in an approximate 50% dilution of candidate cells available for GFP marking. Following reconstitution, approximately 50% of host peripheral blood cells were from the donor (CD45.1+), and out of this population GFP marking remained high 5 months post transplantation in T lymphocytes, myeloid cells (Gr-1/Mac-1), nucleated erythroid cells (Ter119) and B lymphocytes (B220). This percentage of marking was even higher in reconstitution experiments performed without the addition of non-transduced supportive bone marrow, resulting in persistent GFP labeling as high as 98% of cells in the peripheral blood of reconstituted recipients (see the lower left panel of Figure 6D and Supp Figure 8C). The CD45 promoter was also able to support erythroid (Ter119), myeloid (Gr-1/Mac-1), and lymphoid (B220/CD3ε) lineage marking in the bone marrow, spleen and thymus as indicated in Figure 7, confirming that CD45 promoter elements contained within our reporter are sufficient to reliably mark hematopoiesis in lymphoid, myeloid and nucleated erythroid blood cells. To show long term marking, CD45 reporter transduced bone marrow from primary recipients was harvested and transplanted into sublethally irradiated secondary recipients. Again, significant reconstitution was achieved with GFP levels remaining high (95-98%) for up to 22 weeks post-transplant (Figure 7D and Supp Figure 9). This result demonstrates the utility of the CD45 reporting system to serve as an efficient method for documenting stable long-term hematopoietic specification in vivo.
Figure 6. CD45 reporter marks persistent hematopoiesis following transplantation.
(A): Transplantation scheme showing HSPCs derived from CD45.1 strain being transduced with a CD45 reporter followed by transplantation into irradiated congenic CD45.2 recipients. (B): Peripheral blood stained for CD45.1 to determine chimerism in recipient mice that received CD45.1 HSPCs from mock transduced (n=5) and CD45-GFP (n=5) lentiviral reporter constructs. Error bars represent standard deviation. *** represents a statically significant 2 way ANOVA test with p<0.001. (C): Peripheral blood staining at 20 weeks with antibodies detecting CD3ε (T lymphocytes), Gr-1/Mac-1 (granulocytes and macrophages), Ter119 (erythroid progenitors), and B220 (B lymphocytes). Approximately 50% chimerism resulted following transplantation, and the first panel represents donor (CD45.1) cells. The fraction of GFP positivity for each lineage derived from CD45.1 HSPCs is indicated in the pink square box. The positive population for CD45.1 was further sub-gated to determine the percentage of cells representing myeloid and lymphoid lineages. (D): Same as in (C) except following HSPC transplantation without non-transduced supportive bone marrow. Abbreviation: ANOVA, analysis of variance between groups; HSPC, hematopoietic stem and progenitor cell.
Figure 7. CD45 reporter marks blood cell formation in bone marrow and secondary hematopoietic organs, and persists in secondary transplants.
(A): Surface staining on cells of the spleen, (B): thymus, and (C): bone marrow 22 weeks following secondary transplantation. At this time point over 80% of cells come from the donor (CD45.1). High frequency, multilineage marking with the CD45 reporter can be observed. (D) Bone marrow of primary transplant recipient mice was transplanted into secondary CD45.2 recipients. Peripheral blood staining at three weeks post-transplant time shows reconstitution with approximately 95% of CD45.1 cells exhibiting GFP positivity at 22 weeks post-transplantation.
Discussion
All other direct reprogramming platforms have relied on a GFP reporter to assess cellular respecification, clonality, and the efficiency of reprogramming (8, 9, 39), corroborating the utility of this strategy for blood cell derivation. We considered Sca-1 and CD34 gene regulatory elements (unpublished results) as the foundation for our reporter, but they were either not tissue specific, were not expressed in all blood cell lineages (42), or lacked marking strength. In contrast, CD45 was exquisitely restricted to blood cell lineages (Figure 1B and reference 24), making it the most viable candidate that we investigated.
Here we refined efforts from previous studies (26, 27) to locate critical CD45 regulatory elements and identified a 300bp region based on binding signatures for TBP and the blood cell specific transcription factors PU.1, GATA1, c-Myb, MafK and Mxi1. Our results showed that constructs spanning from −164 to +99 of the promoter region drove the highest GFP levels in murine and human cell lines (Figures 2 and 3); this confirms regulatory element activity within this gene promoter is well conserved in mammals. PU.1, Myb, CEBPα/β and GATA binding footprints are required for maximal marking of hematopoietic cells in vivo, supporting the contribution of these critical transcription factors for blood cell specification. This is the first analysis to employ ChIP-seq and DNase-seq for fine mapping and identifying DNA elements sufficient for a lineage specific reporter. Importantly, our reporter is small enough that it can be delivered by traditional lentiviral gene transfer or other genome editing approaches (24, 40).
Using ubiquitous promoters and a high MOI have been fairly effective (25-86% marking) in transducing blood cells in the past (35, 41). However, our blood cell restricted system completely marked the hematopoietic compartment when no additional non-transduced supportive bone marrow was given with the HSPC transplants (98%; see Figure 6D, Supp Figure 8C and Supp Figure 9). We also see high level marking in the spleen, thymus, and bone marrow. Our in vivo analyses demonstrated that the lentiviral CD45 reporter marked blood cells persistently without silencing, evidenced by the robust staining of hematopoietic cells 22 weeks post-secondary transplantation. This has been accomplished using a methodology that results in a significantly lower viral load (24). Importantly, the CD45 reporter is not active in ES cells, and is only activated after these cells had become blood cells (Figure 5). We acknowledge that in transplants receiving only transduced cells there was a minority of cells that did not express GFP. We surmise this is a result of incomplete transduction of the HSPC pool due to our low MOI since the marking levels we observed were persistent across early and late timepoints post-transplantation. The marking efficiency of 98% in one transplant (Figure 6D) implies that silencing, if observed, is not a predominant phenomenon of our lentiviral reporter. Our results suggest that the CD45 reporter system can be used monitor the efficiency of blood cell differentiation from pluripotent cells, to exclude undifferentiated cells from hematopoietic engraftment, and to serve as a simple strategy to follow blood cell transplantation kinetics and persistence via bioluminescence by tethering CD45 regulatory elements to luciferase (42, 43). The reporter can also facilitate tracking of normal and malignant lymphoid cells to interrogate the effects of a genetic loss or gain of function in mouse models following adoptive transfer (44). Additionally, TALEN or CRISPR-Cas9 (40) nucleases can be used to introduce this reporter, together with polycistronic reprogramming vectors, into genomic safe harbors (45). Finally, hematopoietic marking can be coupled with a potent suicide gene we have used previously (46), when driven by a pluripotency gene promoter, to selectively destroy undifferentiated cells and further augment levels of biosafety. With the recent derivation of naïve pluripotent human ES and iPS cell lines that exhibit characteristics of ground state pluripotency (47), it should now be more feasible to generate functional adult hematopoietic cells for the treatment of human blood diseases using this CD45-based GFP marking system.
Supplementary Material
Acknowledgements
We thank the staff of the Carver College of Medicine Flow Cytometry Facility, especially Justin Fishbaugh. We are grateful to Stuart Orkin and Alan Cantor for sharing the MEL cell line, and Gail Bishop for providing the CH12.LX B cell line. This work was supported by startup funds from the Department of Internal Medicine.
This work was supported by startup funds from the Department of Internal Medicine, and University of Iowa Graduate College.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Support and Financial Disclosure Declaration
Conflict of interest disclosure: the authors declare no competing financial interests
Authorship and Disclosures
Conception and design: DL, KD. Collection, assembly of data, data analysis and interpretation: KD, SD, SY, JB, SJ, EK, JC, NZ, HX, DL. Manuscript writing: KD, DL. Providing protocols and reagents: JB, JC, SY, HX, EK, NZ. Final approval of manuscript: DNL. None of the authors declare any competing financial interests.
References
- 1.Shizuru JA, Negrin RS, Weissman IL. Hematopoietic stem and progenitor cells: clinical and preclinical regeneration of the hematolymphoid system. Annual review of medicine. 2005;56:509–38. doi: 10.1146/annurev.med.54.101601.152334. [DOI] [PubMed] [Google Scholar]
- 2.Kaufman DS, Hanson ET, Lewis RL, Auerbach R, Thomson JA. Hematopoietic colony-forming cells derived from human embryonic stem cells. Proc Natl Acad Sci U S A. 2001 Sep 11;98(19):10716–21. doi: 10.1073/pnas.191362598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kyba M, Perlingeiro RC, Daley GQ. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002 Apr 5;109(1):29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- 4.Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007 Dec 21;318(5858):1920–3. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 5.Ran D, Shia WJ, Lo MC, Fan JB, Knorr DA, Ferrell PI, et al. RUNX1a enhances hematopoietic lineage commitment from human embryonic stem cells and inducible pluripotent stem cells. Blood. 2013 Jan 31; doi: 10.1182/blood-2012-08-451641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szabo E, Rampalli S, Risueno RM, Schnerch A, Mitchell R, Fiebig-Comyn A, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010 Nov 25;468(7323):521–6. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 7.Pereira CF, Chang B, Qiu J, Niu X, Papatsenko D, Hendry CE, et al. Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell. 2013 Aug 1;13(2):205–18. doi: 10.1016/j.stem.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010 Feb 25;463(7284):1035–41. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010 Aug 6;142(3):375–86. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012 Jan 19;481(7381):295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol. 2010 May;28(5):521–6. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009 Nov;27(11):1033–7. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 13.Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010 Jul 2;7(1):64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 14.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008 Mar 6;2(3):230–40. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013 Aug 9;341(6146):651–4. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- 16.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008 Feb 22;132(4):631–44. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossi L, Lin KK, Boles NC, Yang L, King KY, Jeong M, et al. Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell. 2012 Sep 7;11(3):302–17. doi: 10.1016/j.stem.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki N, Ohneda O, Minegishi N, Nishikawa M, Ohta T, Takahashi S, et al. Combinatorial Gata2 and Sca1 expression defines hematopoietic stem cells in the bone marrow niche. Proc Natl Acad Sci U S A. 2006 Feb 14;103(7):2202–7. doi: 10.1073/pnas.0508928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hills D, Gribi R, Ure J, Buza-Vidas N, Luc S, Jacobsen SE, et al. Hoxb4-YFP reporter mouse model: a novel tool for tracking HSC development and studying the role of Hoxb4 in hematopoiesis. Blood. 2011 Mar 31;117(13):3521–8. doi: 10.1182/blood-2009-12-253989. [DOI] [PubMed] [Google Scholar]
- 20.Kataoka K, Sato T, Yoshimi A, Goyama S, Tsuruta T, Kobayashi H, et al. Evi1 is essential for hematopoietic stem cell self-renewal, and its expression marks hematopoietic cells with long-term multilineage repopulating activity. J Exp Med. 2011 Nov 21;208(12):2403–16. doi: 10.1084/jem.20110447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y, Lim KC, Onodera K, Takahashi S, Ohta J, Minegishi N, et al. Rescue of the embryonic lethal hematopoietic defect reveals a critical role for GATA-2 in urogenital development. EMBO J. 1998 Nov 16;17(22):6689–700. doi: 10.1093/emboj/17.22.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. P Natl Acad Sci USA. 2004 Apr 20;101(16):6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma X, Robin C, Ottersbach K, Dzierzak E. The Ly-6A (Sca-1) GFP transgene is expressed in all adult mouse hematopoietic stem cells. Stem Cells. 2002;20(6):514–21. doi: 10.1634/stemcells.20-6-514. [DOI] [PubMed] [Google Scholar]
- 24.Levasseur DN, Ryan TM, Pawlik KM, Townes TM. Correction of a mouse model of sickle cell disease: lentiviral/antisickling beta-globin gene transduction of unmobilized, purified hematopoietic stem cells. Blood. 2003 Dec 15;102(13):4312–9. doi: 10.1182/blood-2003-04-1251. [DOI] [PubMed] [Google Scholar]
- 25.Hermiston ML, Xu Z, Weiss A. CD45: a critical regulator of signaling thresholds in immune cells. Annual review of immunology. 2003;21:107–37. doi: 10.1146/annurev.immunol.21.120601.140946. [DOI] [PubMed] [Google Scholar]
- 26.Timon M, Beverley PC. Structural and functional analysis of the human CD45 gene (PTPRC) upstream region: evidence for a functional promoter within the first intron of the gene. Immunology. 2001 Feb;102(2):180–9. doi: 10.1046/j.1365-2567.2001.01177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anson DS, Occhiodoro T. Transcriptional activity of the CD45 gene promoter in retroviral vector constructs. Biochim Biophys Acta. 1994 Sep 13;1219(1):81–8. doi: 10.1016/0167-4781(94)90249-6. [DOI] [PubMed] [Google Scholar]
- 28.Virts EL, Raschke WC. The role of intron sequences in high level expression from CD45 cDNA constructs. J Biol Chem. 2001 Jun 8;276(23):19913–20. doi: 10.1074/jbc.M100448200. [DOI] [PubMed] [Google Scholar]
- 29.Yang J, Hills D, Taylor E, Pfeffer K, Ure J, Medvinsky A. Transgenic tools for analysis of the haematopoietic system: knock-in CD45 reporter and deletor mice. Journal of immunological methods. 2008 Sep 15;337(2):81–7. doi: 10.1016/j.jim.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 30.Chan KM, Bonde S, Klump H, Zavazava N. Hematopoiesis and immunity of HOXB4-transduced embryonic stem cell-derived hematopoietic progenitor cells. Blood. 2008 Mar 15;111(6):2953–61. doi: 10.1182/blood-2007-10-117366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKinney-Freeman SL, Naveiras O, Daley GQ. Isolation of hematopoietic stem cells from mouse embryonic stem cells. Curr Protoc Stem Cell Biol. 2008 Feb; doi: 10.1002/9780470151808.sc01f03s4. Chapter 1:Unit 1F 3. [DOI] [PubMed] [Google Scholar]
- 32.Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, et al. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome research. 2008;4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas ML. The leukocyte common antigen family. Annual review of immunology. 1989;7:339–69. doi: 10.1146/annurev.iy.07.040189.002011. [DOI] [PubMed] [Google Scholar]
- 34.Hao QL, Shah AJ, Thiemann FT, Smogorzewska EM, Crooks GM. A functional comparison of CD34 + CD38− cells in cord blood and bone marrow. Blood. 1995 Nov 15;86(10):3745–53. [PubMed] [Google Scholar]
- 35.Chen W, Wu X, Levasseur DN, Liu H, Lai L, Kappes JC, et al. Lentiviral vector transduction of hematopoietic stem cells that mediate long-term reconstitution of lethally irradiated mice. Stem Cells. 2000;18(5):352–9. doi: 10.1634/stemcells.18-5-352. [DOI] [PubMed] [Google Scholar]
- 36.Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010 Oct 8;7(4):532–44. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 37.Whitlock CA, Witte ON. Long-term culture of B lymphocytes and their precursors from murine bone marrow. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3608–12. doi: 10.1073/pnas.79.11.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sudo T, Ito M, Ogawa Y, Iizuka M, Kodama H, Kunisada T, et al. Interleukin 7 production and function in stromal cell-dependent B cell development. J Exp Med. 1989 Jul 1;170(1):333–8. doi: 10.1084/jem.170.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011 Jul 21;475(7356):386–9. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 40.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013 Sep 12;154(6):1380–9. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mostoslavsky G, Kotton DN, Fabian AJ, Gray JT, Lee JS, Mulligan RC. Efficiency of transduction of highly purified murine hematopoietic stem cells by lentiviral and oncoretroviral vectors under conditions of minimal in vitro manipulation. Mol Ther. 2005 Jun;11(6):932–40. doi: 10.1016/j.ymthe.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 42.Wang X, Rosol M, Ge S, Peterson D, McNamara G, Pollack H, et al. Dynamic tracking of human hematopoietic stem cell engraftment using in vivo bioluminescence imaging. Blood. 2003 Nov 15;102(10):3478–82. doi: 10.1182/blood-2003-05-1432. [DOI] [PubMed] [Google Scholar]
- 43.Tian X, Hexum MK, Penchev VR, Taylor RJ, Shultz LD, Kaufman DS. Bioluminescent imaging demonstrates that transplanted human embryonic stem cell-derived CD34(+) cells preferentially develop into endothelial cells. Stem Cells. 2009 Nov;27(11):2675–85. doi: 10.1002/stem.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu S, Zhou X, Steinke FC, Liu C, Chen SC, Zagorodna O, et al. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 2012 Nov 16;37(5):813–26. doi: 10.1016/j.immuni.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sadelain M, Papapetrou EP, Bushman FD. Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer. 2012 Jan;12(1):51–8. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 46.Hong JS, Waud WR, Levasseur DN, Townes TM, Wen H, McPherson SA, et al. Excellent in vivo bystander activity of fludarabine phosphate against human glioma xenografts that express the escherichia coli purine nucleoside phosphorylase gene. Cancer Res. 2004 Sep 15;64(18):6610–5. doi: 10.1158/0008-5472.CAN-04-0012. [DOI] [PubMed] [Google Scholar]
- 47.Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013 Dec 12;504(7479):282–6. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.