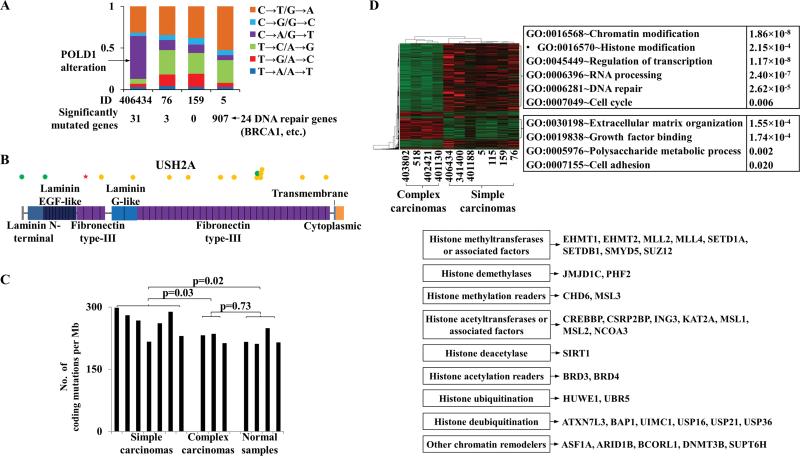
Figure 3. Coding sequence mutations are frequent in canine simple carcinomas; chromatin-modification genes are downregulated in canine complex carcinomas.
A, the fractions (the Y-axis) of somatic base substitution types of simple carcinomas (IDs indicated by the X-axis) detected by WES. The total number of significantly mutated genes in each tumor is also shown, and tumor 5 has many DNA repair genes mutated.
B, synonymous (green dots) and non-synonymous substitutions (yellow dots), and a nonsense mutation (red star) uncovered in the USH2A gene in tumor 5.
C, the base substitution (compared to the dog reference genome) rates of the three sample types in coding regions with 30-300X RNA-seq read coverage. The p-values were calculated by t-tests.
D, the heatmap of 751 genes differentially expressed at FDR ≤ 0.2 between simple and complex carcinomas (red: upregulation; green: downregulation). The right panel illustrates the enriched functions of each gene cluster indicated, and the 35 chromatin modifiers downregulated in complex carcinomas are specified below.