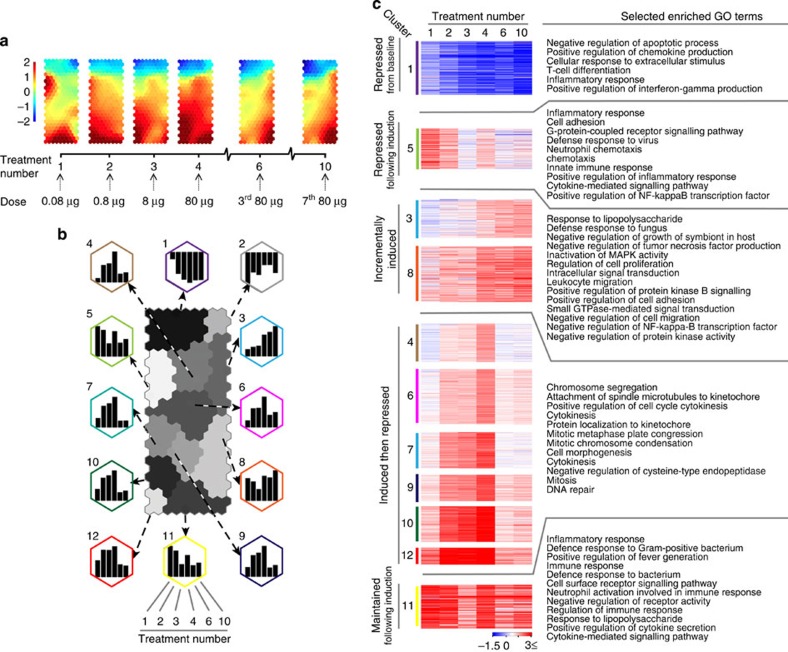
Figure 6. Transcriptome analysis of CD4+ T cells at consecutive stages of escalating dose immunotherapy.
Tg4 Rag-1−/−mice were EDI-treated s.c. with MBP Ac1-9[4Y]. CD4+ T-cell transcriptome analysis was undertaken at the indicated treatment stages (n=3 per time point, RNA pooled). (a) CPP-SOM of 1,893 regulated transcripts across six stages of EDI, illustrating treatment stage-specific transcriptional changes, relative to PBS-treated controls. Hexagons represent groups of co-clustered transcripts; colour changes show modulation of transcript expression (upregulation in red, downregulation in blue and moderate regulation in yellow and green). (b) Twelve gene clusters (colour-coded, labelled 1–12) obtained by two-phase SOM clustering. Bar graphs show the expression pattern of the seed unit used to derive each gene cluster. (c) Heatmaps showing expression patterns of clustered genes, and GO terms associated with each. Gene clusters are organized according to five dominant patterns; genes that are repressed from baseline, repressed following initial induction, incrementally induced, induced and then repressed and finally, induced and then maintained throughout the course of treatment. All GO terms are associated with at least three transcripts within a cluster, with a false-discovery rate (FDR) of <0.05.