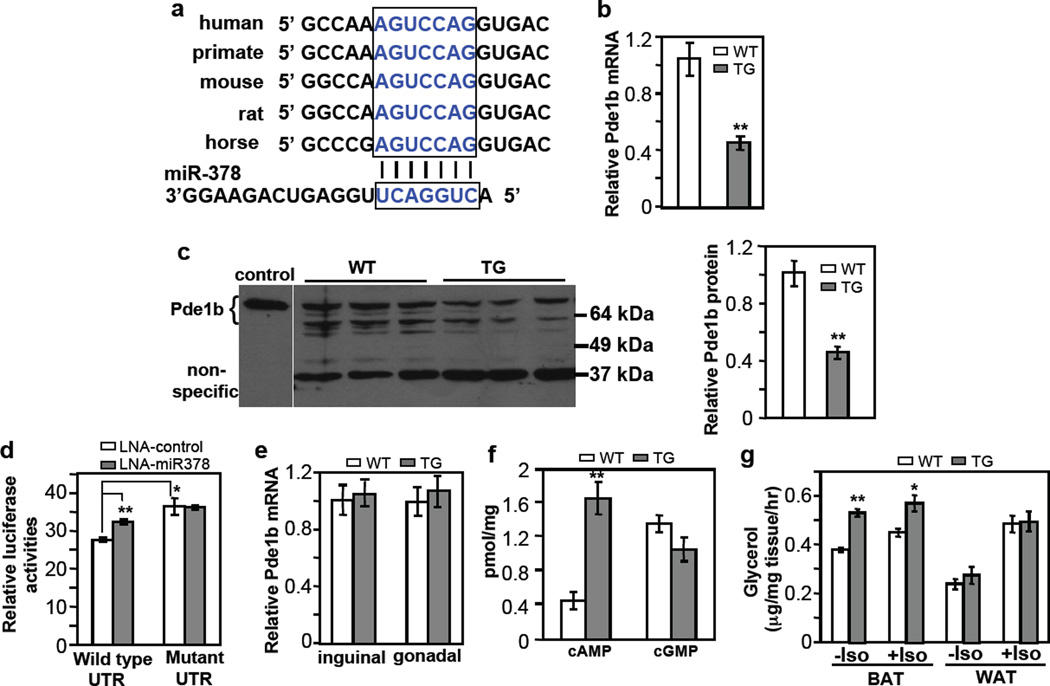
Fig. 6. BAT-specific suppression of Pde1b by miR-378.
(a) Conserved miR-378 targeting sequence at Pde1b 3’ UTR.
(b) Pde1b mRNA is downregulated in BAT of miR-378 transgenic mice. n=5 mice per group (9-week-old male mice), mean ± SE, **P<0.001 (two-tailed Student’s t-test).
(c) Pde1b protein levels in BAT from 5-week-old female WT and TG mice. Cell extracts from HEK293 cells transfected with a full-length Pde1b1 plasmid were used as a control to indicate the position of endogenous mouse Pde1b. BAT produces two expected major splice variants of Pde1b (Pde1b1 and Pde1b2) differing in their amino-termini70. Right panel shows quantification of the western blot. Mean ± SE, **P=0.005 (two-tailed Student’s t-test).
(d) HEK293 cells were transfected with locked nucleic acid (LNA) miR-378 inhibitor and a luciferase reporter carrying 3’ UTR of Pde1b as described in the online methods. Luciferase activity was measured. n=3, mean ± SE, *P<0.05, **P<0.001 (two-tailed Student’s t-test).
(e) Relative Pde1b mRNA level in inguinal and gonadal WAT of the transgenic mice. n=5 mice per group (9-week-old male mice), mean ± SE.
(f) cAMP and cGMP levels in BAT. n=7 mice per group (9-week-old male mice), mean ± SE, **P<0.001 (two-tailed Student’s t-test).
(g) BAT and WAT lipolysis with or without 10 µM isoproterenol. n=5 mice per group (5-month-old male mice), mean ± SE, **P<0.0005, *P=0.012 (two-tailed Student’s t-test).