Figure 1.
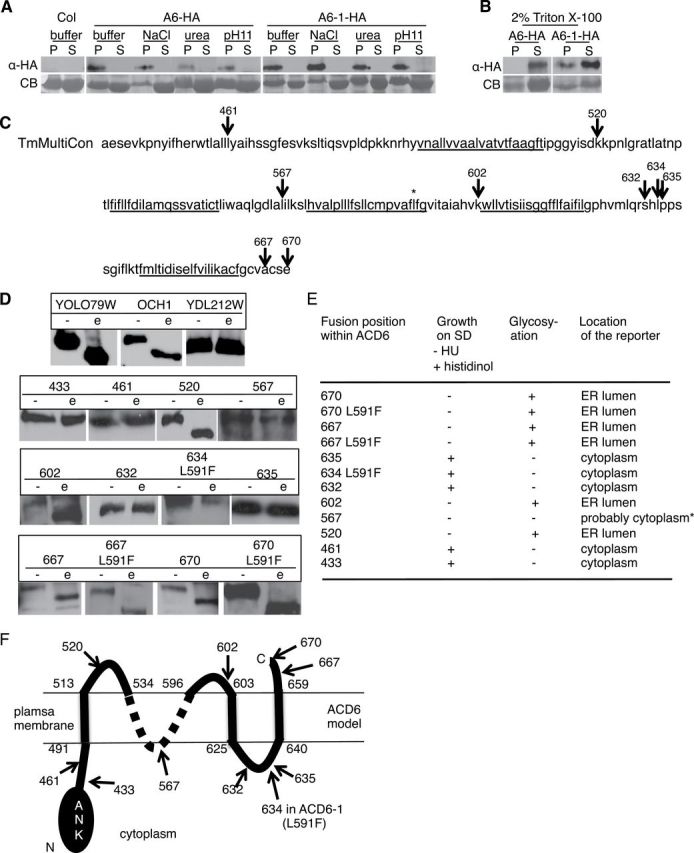
ACD6 and ACD6-1 Are Multipass Integral Membrane Proteins.
(A, B) ACD6-HA and ACD6-1-HA are integral membrane proteins (A) that can be solubilized with detergent (B). Microsomal protein from transgenic plants expressing ACD6-HA (A6-HA)/ACD6-1-HA (A6-1-HA) treated with 1.5M NaCl, 2M urea, 100mM Na2CO3 (pH 11), or 2% Triton X-100, respectively, partitioned into supernatant (S) or membrane pellet (P) fractions and concentrated using trichloroacetic acid were subjected to immunoblotting using HA antibody. CB, Coomassie blue-stained membrane. Experiments in (A) and (B) were repeated twice, with similar results.
(C) A consensus model from the Aramemnon database showing potential membrane spanning regions underlined within the C-terminus of ACD6 (amino acids 441–670). * marks the L591F change in ACD6-1.
(D) ACD6 truncation fusion proteins produced in yeast were treated with endo-glycosidase H (e) or mock-treated (–) and subject to Western blot analysis using HA antibody. Top panel shows yeast positive (YOLO79W and OCH1) and negative (YCL212W) control proteins for deglycosylation.
(E) Summary of the growth of yeast producing the fusion proteins and predicted location of the C-terminus of each fusion protein based on results from (D) and the growth assays. For the growth assay, if the C-terminus of the fusion protein is in the cytosol and folds properly, yeast grows on histidinol. Localization to the ER lumen (evidenced by glycosylation of the reporter fusion partner) predicts an extracellular location in plant cells. The lack of growth of fusion 567 (*) on histidinol was likely due to the lack of productive folding of the fusion partner.
(F) Provisional topology model for ACD6. Numbers demarcating the extent of transmembrane regions are inferred from the ARAMEMNON TmMultiCon analysis and our data. The ambiguity of the structure in the 536–594 region is denoted by a dotted line. Given the predicted topology of ACD6 (panel (C)), its likely that 567 is in a cytosolic loop. We were unable to resolve the ambiguity of the location of the region near 567 with additional fusions. At least two independent yeast transformants per construct were used for each assay. Numbers (C), (D), and (F) mark the junctions of C-terminal reporter.
