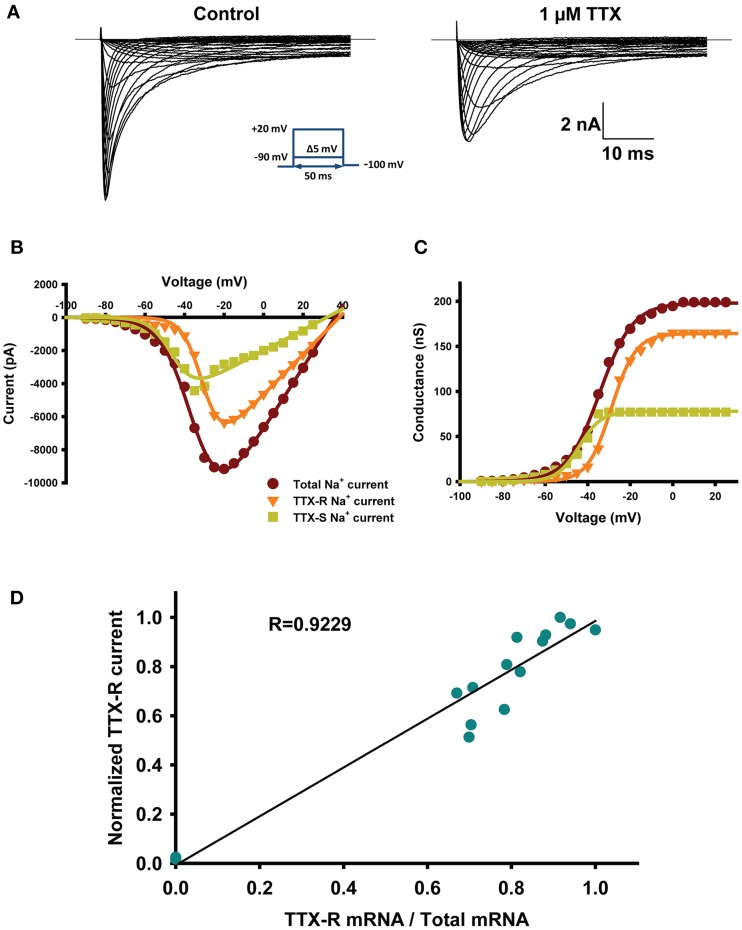
Figure 1.
mRNAs reflect electrophysiological properties. (A) Representative current-traces recorded from small diameter DRG neurons (28 μm in diameter) before (right) and after (left) the addition of 1 μM TTX. The protocol is shown in the inset. (B) Example of current-voltage (I–V) relationships obtained from the small diameter DRG neuron shown in (A). The dark red trace is the total current recorded under control conditions, the orange trace is the total TTX-R current recorded in presence of 1 μM TTX, and the green trace is the TTX-S current obtained by subtracting the TTX-R Nav current (orange) from the total Nav current (dark red). I–V are fitted using a Hodgkin–Huxley-like equation: f = Gmax*(V − Vrev)/ (1 + exp((V − V1/2)/k)), where Gmax is the maximal conductance, V the potential, V1/2 is the voltage at which half of the channels are in the open state, Vrev is the reversal potential, and k is the slope factor. (C) Activation curves obtained from (B) illustrating the conductance of total Nav current (dark red), TTX-R current (orange) and TTX-S current (green). The conductance was calculated using the following equation: GNa = INa/(Vm − Vrev), where GNa is the conductance, INa is the peak current for the test potential Vm, and Vrev is the reversal potential estimated from the current-voltage curve. (D) Correlation of normalized TTX-R Nav currents to normalized mRNA coding for TTX-R channels (Nav1.8 + Nav1.9/(Nav1.7 + Nav1.8 + Nav1.9)). Currents were normalized to the maximal current. n = 17 from 8 animals.