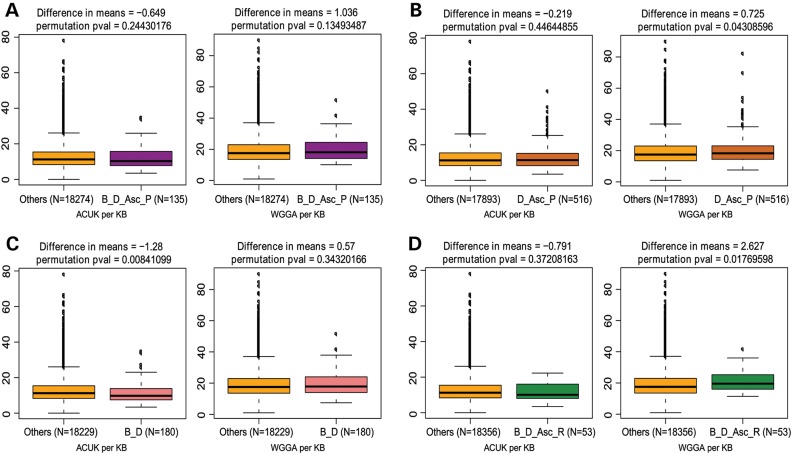
Figure 4.
Frequency of ACUK and WGGA among the Consensus FMRP targets. Sequence motif frequency evaluation of the genes common to the Brown, Darnell, Ascano PAR-CLIP or RIP-Chip data. The box plots show the median, quartiles, 1.5× interquartile range, and outliers in the number of ACUK (left) and WGGA (right) patterns per kilobase compared with the rest of the genes in the genome (Others). The difference in the mean number of patterns per kilobase between the two groupings and the permutation P-value are given at the top of each graph; the number of genes in each grouping is given below each graph; number of permutations = 1 000 000. The different dataset combinations are as follows: (A) Ascano PAR-CLIP, Brown and Darnell datasets; (B) Ascano PAR-CLIP and Darnell; (C) Brown and Darnell; and (D) Ascano RIP-Chip, Brown and Darnell datasets. B, Brown; D, Darnell; Asc-P, Ascano PAR-CLIP; Asc-R, Ascano RIP-Chip.