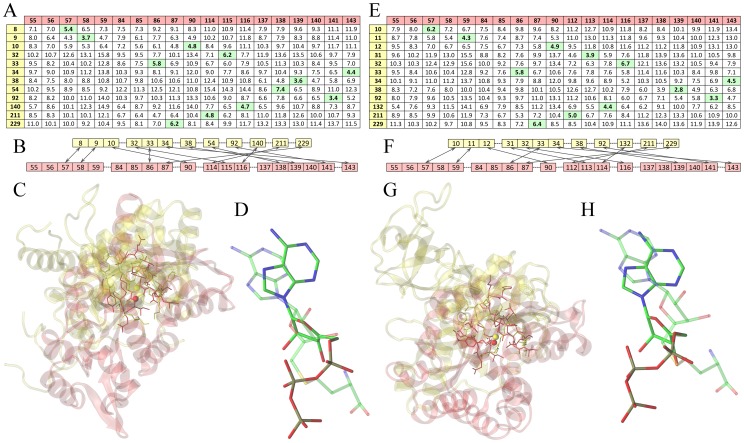
Figure 3. Construction of sequence order-independent binding site alignments by eMatchSite.
Two target proteins are ATP-dependent DNA ligase (PDB-ID: 1a0iA, yellow) and histamine N-methyltransferase (PDB-ID: 2aotA, red). Left (A–D) and right (E–H) panels show the alignment of binding sites in the crystal structures and protein models, respectively. (A, E) Matrices of pairwise Cα-Cα distances between two binding sites predicted by SVR. Residue indexes are shown in the first column and row. Sets of residue pairs that have the smallest Cα-Cα distances identified by the Kuhn-Munkres algorithm are highlighted in green. (B, F) Sequence order-independent alignments of two binding sites constructed from residue pairs that have the smallest Cα-Cα distances; arrows indicate equivalent pairs. (C, G) Protein structures are superposed according to the local alignment of their binding sites; binding residues and predicted pocket centers are shown as solid sticks and balls, respectively. (D, H) Relative orientation of binding ligands upon the local alignment of target binding sites; ATP in 1a0iA and S-adenosyl-L-homocysteine in 2aotA are shown as solid and transparent sticks, respectively.