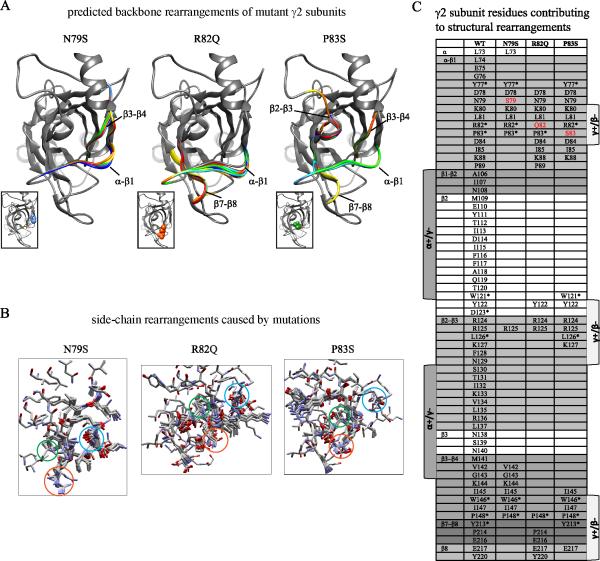
Figure 8. Structural simulation predicted mutation-induced changes in subunit structure.
A. Superpositions of structural models for up to 10 of the best-scoring low-energy generated backbones of wildtype and mutated N79S, R82Q and P83S γ2 subunits were made. In ribbon representation, the native secondary structure was shown in gray, and the mutated secondary structures were represented by other colors. Structural domains as shown in panel C were also represented. Sites of missense mutations in γ2 subunits were shown in space-filling representation in the inserts: N79 in blue, R82 in orange, and P83 in green. B. Local side-chain rearrangements observed for mutated N79S, R82Q and P83S γ2 subunit residues were displayed. Neighboring residues within an 11 Å radius were shown in stick representation and color by element (CPK representation). Sites of wildtype or mutated N79S, R82Q or P83S residues were labeled in blue, orange or green circles. C. A table of predicted amino acids contributing to side-chain rearrangements for mutated residues at positions 79, 82 and 83 was categorized by structural domains. The mutated residues were shown in red, and identical residues among the Cys-loop family were labeled with an asterisk.