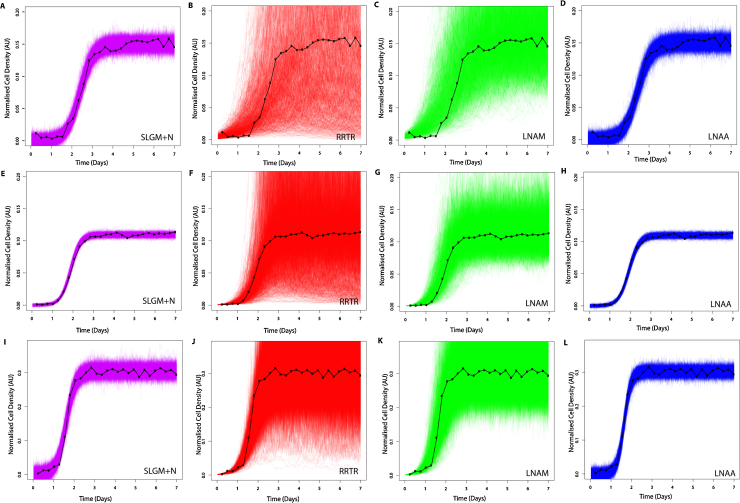
Fig. 3.
Forward trajectories with measurement error, simulated from inferred parameter posterior samples (sample size = 1000). Model fitting is carried out on SLGM forward trajectories with normal measurement error (black), for three different sets of parameters (see Table 2). See (16) or (17) for model and Table C.1 for prior hyper-parameter values. Each row of figures corresponds to a different time course data set, simulated from a different set of parameter values, see Table 2. Each column of figures corresponds to a different model fit: (A), (E) and (I) SLGM+N (pink). (B), (F) and (J) RRTR model with lognormal error (red). (C), (G) and (K) LNAM model with lognormal error (green). (D), (H) and (L) LNAA model with normal error (blue). See Table 2 for parameter posterior means and true values.(For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)