Figure 2.
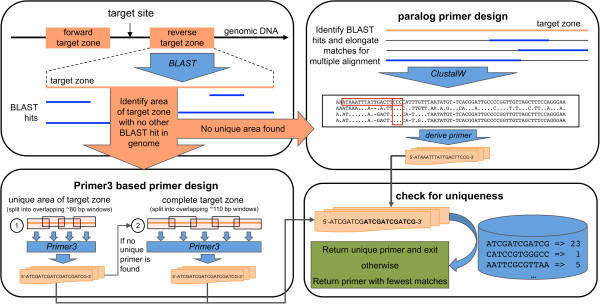
Overview on the primer design tool. Initially, each of the two target zones with a distance to the chosen target site (distance and target site defined by the user) is examined by a BLAST vs. the A. thaliana genome sequence. If there is an area that has no other BLAST hit somewhere in the genome, the Primer3-based approach is used (bottom left box), otherwise the paralog primer design is used (upper right box). A number of candidate primers is designed in both approaches which are then checked for uniqueness (bottom right box). If a unique primer with no additional 12 bp-hit at the 3′-end in the genome is found, the primer design is stopped and the primer is returned as a result. Otherwise the primer with the fewest matches is returned. When designing additional primers, the next best primers are returned. The Primer3-based primer design (bottom left box) uses multiple runs of Primer3 in overlapping windows and altering temperatures to generate a large set of candidate primers. First, only the unique area of the target zone with no additional BLAST hit in the genome is considered. If no unique primer is found, the process is performed again with the complete target zone. The paralog primer design first creates a multiple alignment with all sequences showing a BLAST hit to the target zone using ClustalW. The algorithm searches for mismatches in the multiple alignment (see Figure 3 for details). To reduce the runtime of ClustalW for sequences with many hits, the target zone is also split into overlapping windows and alignments are computed separately (not shown in Figure). The primer sequences shown in the figure are to be regarded as example sequences that cannot fit to *all* features of the scheduled workflow.
