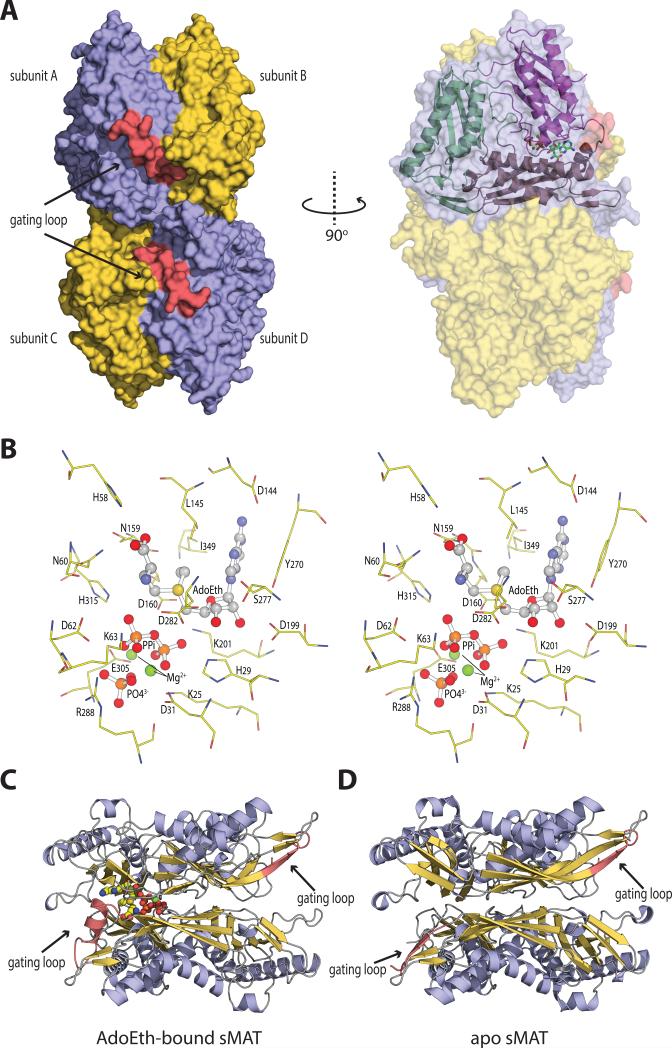
Fig. 3. The overall molecular structure and active site contents of sMAT.
(A) Tetramer assembly of sMAT in the crystal structure as calculated by PISA. The surface of the protein is displayed: four protein monomers are shown in light blue or yellow and the gating loops are shown in red. One protein monomer is displayed on the right showing the three intertwined domains as cartoon and the ligands as sticks.
(B) Stereoview of sMAT-ligands interactions. The stick model of AdoEth, PPi, PO43- and Mg2+ is depicted in spheres and the interacting sMAT residues are labeled and illustrated in green.
(C) Side view of sMAT dimer with AdoEth bound. The gating loop region is highlighted in red.
(D) Side view of apo sMAT dimer. The gating loop region is highlighted in red.