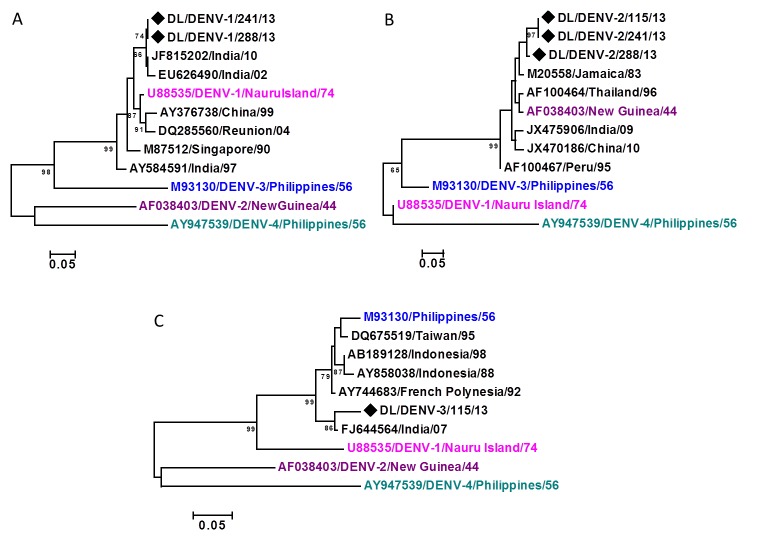
Maximum Likelihood phylogenetic tree based on C-prM gene region of A) DENV-1 strains B) DENV-2 strains C) DENV-3 strain .
The sequences were obtained from co-infecting strains detected in the outbreak. The sequences obtained in the study are marked by black diamonds. Other strains are represented by their GenBank accession number followed by country of origin and last two digits of year of isolation. The numbers on nodes represent bootstrap values generated by 1000 replications. Bootstrap values of >65 are shown. The branch lengths are proportional to the number of nucleotide changes as indicated by the scale bar (0.05 substitutions per site). Prototype strains of all 4 dengue serotypes are included in the trees.