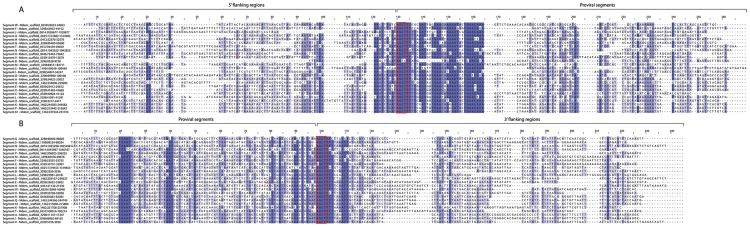
Figure 2. Wasp Integration Motifs (WIMs) for all MdBV proviral segments.
(A) Alignment of 200 nucleotides (nt) surrounding the 5′ WIM site of each segment with similarity for each site colored in shades of blue. Red box highlights the tetramer AGCT. 31 nt are conserved in each proviral segment following the AGCT motif (red box), while the 5′ flanking region upstream of this motif is AT rich and not well conserved. (B) Alignment of 200 nt surrounding the 3′ WIM site for proviral segments shows that the flanking region preceding the WIM site is AT rich but not conserved, whereas the first 100 nt of each proviral segment downstream of the WIM site shows high conservation. Maximum likelihood analyses indicated that the relationships between segments for the WIM sites (A) and 3′ flanking regions (B) cannot be resolved.