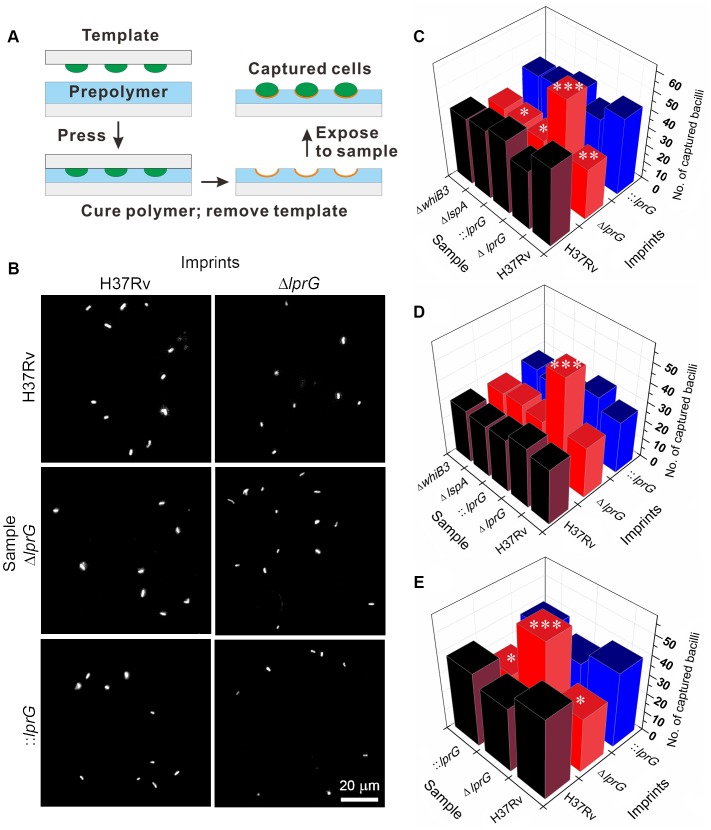
Figure 2. Cell-imprinting reveals altered cell surface and reduced surface-exposed LAM in the lprG mutant.
(A) Illustration of cell-imprinting and cell capture. Equal number of bacteria grown in shaking broth culture with Tween-80 were fixed in paraformaldehyde, washed with PBS, dried on a polystyrene glass slide, and then used as a template for imprinting. The template was pressed into pre-cured PDMS polymer which was then cured at 37°C for 8 h, followed by 60°C for 1 h. The template was peeled off and the imprinted polymer film was sonicated in distilled water for 5 min. Samples of bacteria suspended in PBS at an ODA580 of 0.01 were fluorescently labeled with propidium iodide and 25 µl was flowed at 5 µl/min through a microfluidic device containing the imprints of wild-type (H37Rv), lprG mutant (ΔlprG), and ΔlprG complemented with lprG-Rv1410c (::lprG). The average number of captured bacilli per eight view fields was measured using a fluorescent microscope. (B) Representative images showing captured H37Rv, ΔlprG, and ::lprG on imprints of H37Rv and ΔlprG. (C–E) Bar graphs show the average number of captured H37Rv, ΔlprG, ::lprG, lspA mutant (ΔlspA) and whiB3 mutant (ΔwhiB3). Samples were pretreated with PBS alone (C), with rabbit anti-LAM pAb (α-LAM) at a dilution of 1/20 (D) and with mouse anti-Ag85 complex mAb (CS-90) at a dilution of 1/10 (E). Data is representative of three independent experiments. Capture on imprints of ΔlprG and ::lprG was compared to H37Rv. *P<0.05; **P<0.01; ***P<0.005.