Figure 1.
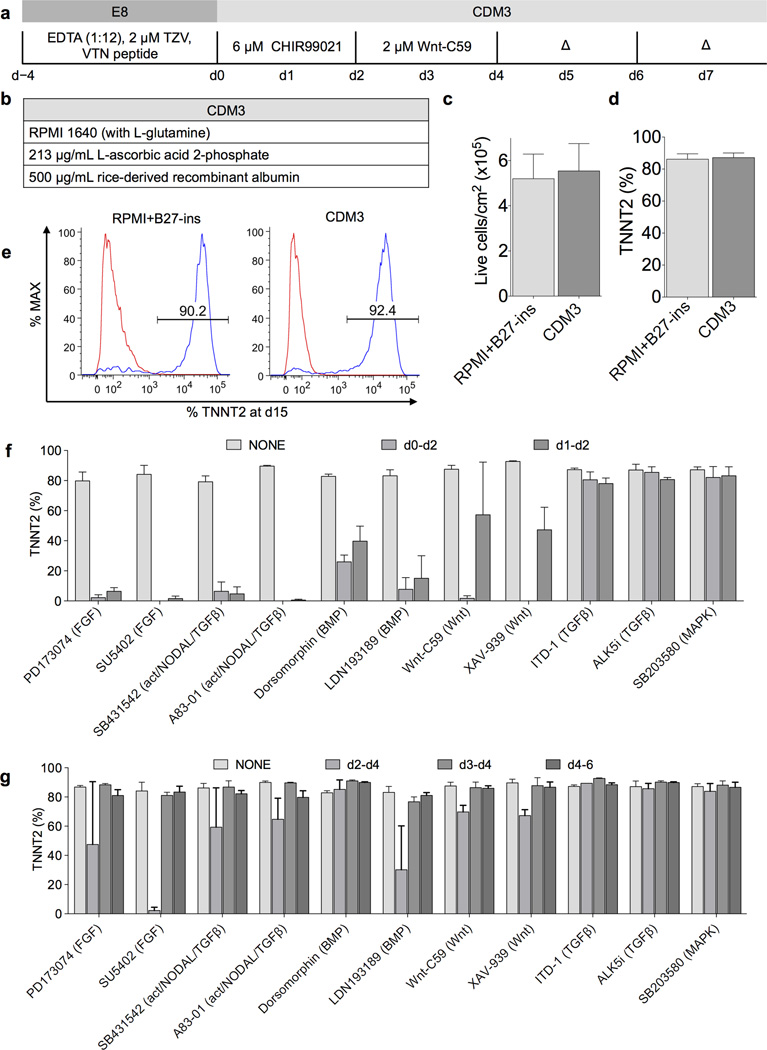
Chemically defined differentiation protocol for efficient cardiac differentiation of hiPSCs. a) Schematic of optimized chemically defined cardiac differentiation protocol. E8, chemically defined pluripotency medium; EDTA, ethylenediaminetetraacetic acid used for clump cell passaging; TZV, thiazovivin, a Rho kinase inhibitor; VTN, vitronectin; CDM3, chemically defined medium 3 components; Δ, medium change. b) Simple three-component formula of CDM3. c) Comparison of total live cell yield from differentiations in RPMI+B27-ins and CDM3, measured by flow cytometry for cardiac troponin T (TNNT2) on day 15 cells (hiPSC line 59FSDNC3 shown), n = 4. d) Comparison of cardiac differentiation efficiency from differentiations in RPMI+B27-ins and CDM3, n = 4. e) Typical TNNT2+ populations in cells produced from differentiations in RPMI+B27-ins medium and CDM3. Left peak represents isotype control. f) Effect of inhibition of signaling pathways during early mesoderm differentiation. Small molecules were added at designated time points (day 0–2 or day 1–2), at 5 µM for all except Wnt-C59, which was used at 2 µM. Normal doses of CHIR99021, Wnt-C59, and medium change timings were maintained, n = 3. g) Effect of inhibition of signaling pathways post mesoderm induction. The same as f) but assessing cardiac induction time points (day 2–4, day 3–4, or day 4–6). Slight reduction of cardiac differentiation efficiency with Wnt-C59 on day 2 to day 4 was likely due to the combined total dose (4 µM) being suboptimal (Supplementary Fig. 4i), n = 3. All error bars represent S.E.M.