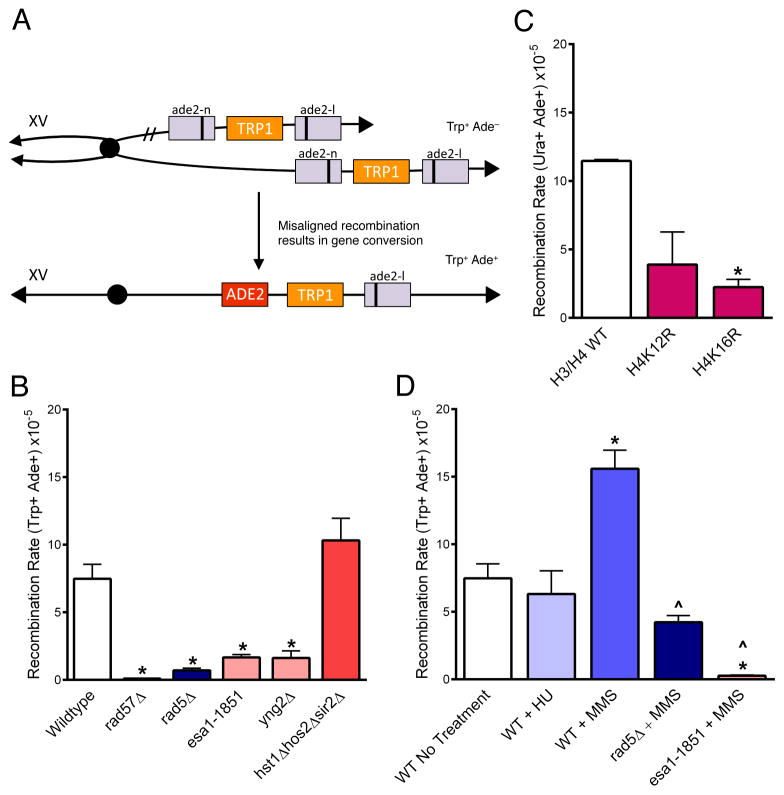
Figure 4. Sister chromatid recombination depends on Esa1, Rad5, and H4K16 acetylation.
(A) Unequal sister chromatid gene conversion from Trp+ Ade− to Trp+ Ade+ can be used as a measure of sister chromatid recombination rates; Ade+ recombinants can also arise via intra-chromatid gene conversion but selection for Trp+ (or Ura+ in (C)) eliminates intrachromatid pop-out recombinants (Mozlin, 2008). Only gene conversion of the lower chromatid and the resulting product is shown. (B) Rates of spontaneous SCR. (C) SCR was measured in H4 point mutants using a Ura+ Ade- construct (URA3 in place of TRP1). Point mutants were expressed from a plasmid in cells in which the endogenous H3 and H4 genes were deleted. Point mutants were compared to a strain that expresses the H3/H4 wildtype from the plasmid for statistical analysis. (D) SCR rates were measured in cells treated for one hour with MMS (0.033%), or HU (0.2M). Statistical deviation using the Student’s t-test is indicated, *p<0.05 from wildtype, ^p<0.05 from wildtype MMS treated. Data are the average of at least 3 experiments +/− SEM. See also Table S3.