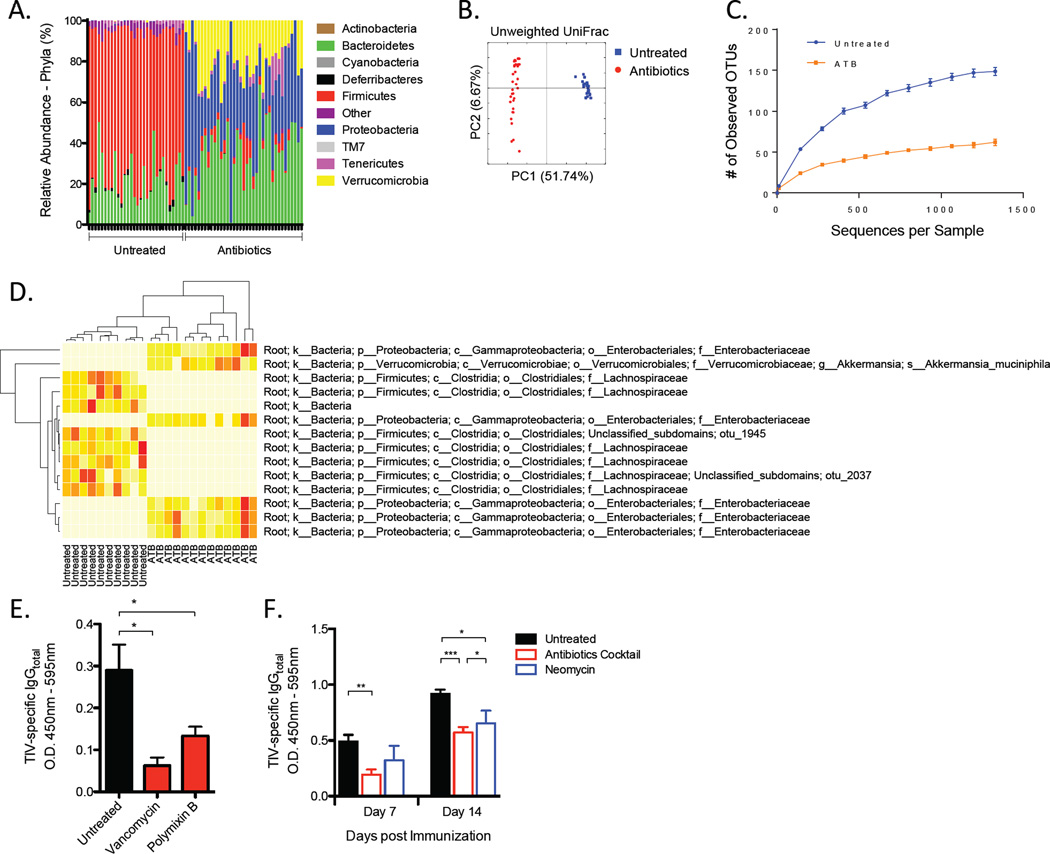
Figure 4. Multiple Types of Bacteria Are Necessary to Mediate Antibody Response to TIV.
(A–D) Composition of the microbiota in antibiotics-treated or untreated mice. Diversity and specific bacterial components of the host microbiota were assessed using genomic bacterial DNA sequence analyses as described in Experimental Procedures.
(A) Relative abundance of specific bacterial phyla present in the two mouse groups.
(B) Unweighted Unifrac principal coordinate analyses using the QIIME analysis software.
(C) Rarefaction curves for untreated or antibiotics treated (ATB) groups generated based on the sequenced 16S rRNA gene libraries. Data are depicted as the number of unique operational taxonomic units (OTUs).
(D) A “nearest-shrunken centroid” classification analyses identifying bacterial taxa groups highly represented in either of the two clustered groups. Data shown are relative abundance of each OTU identified by this classification method represented in a heat map (white (least abundance) to red (most abundant)).
(E) TIV-specific IgG concentrations on day 7-post vaccination in mice untreated or treated with either vancomycin or polymixin B. Raw O.D. values shown were obtained using serum diluted by a factor of 1:200.
(F) TIV-specific IgG concentrations in mice treated with antibiotics or with neomycin alone. Raw O.D. values shown were obtained using serum diluted by a factor of 1:200 and expressed as means ± SEM. See also Figure S4.