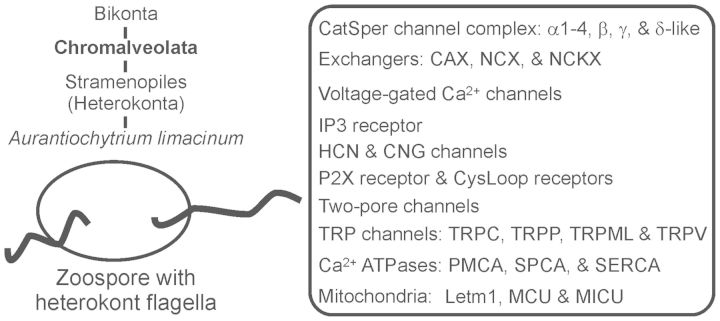
Fig. 2.
Ca2+ signaling machinery in Aurantiochytrium limacinum. The evolutionary relationship of A. limacinum is inferred from the Tree of Life project (http://www.tolweb.org/) and the JGI genome portal (http://genome.jgi-psf.org/Aurli1/Aurli1.home.html, last accessed July 11, 2014). Aurantiochytrium limacinum life stages include colonies of vegetative cells or zoospores with heterokont flagella (http://syst.bio.konan-u.ac.jp/labybase/Aurantiochytrium_limacinum_life_cycle.html, last accessed July 11, 2014). CatSper, sperm-associated cation channel; CNG, cyclic nucleotide-gated channel; CysLoop receptors, cysteine-loop ligand-gated receptor; IP3 receptor, inositol 1,4,5-trisphosphate receptor; Letm1, leucine zipper-EF-hand containing transmembrane protein 1; MCU, mitochondrial Ca2+ uniporter; MICU, mitochondrial EF hand Ca2+ uniporter regulator; NC(K)X, Na+/Ca2+ (K+-dependent) exchanger; P2X receptor, P2X purinergic receptor; PMCA, plasma membrane Ca2+ ATPase; SERCA, sarco/endoplasmic reticulum Ca2+ ATPase; SPCA, secretory pathway Ca2+ ATPase.