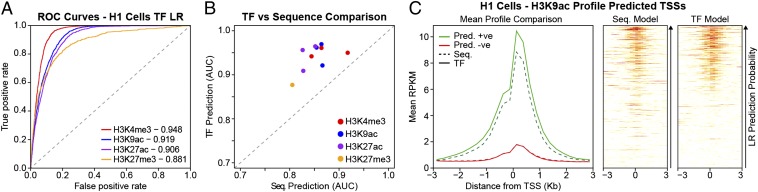
Fig. 2.
Histone modifications can be predicted from TF-binding data. (A) Representative ROC curves of the performance of TF ChIP-Seq LR-based classifiers for histone modifications at gene promoters in H1 cells. The AUC for each task is indicated in the legend. The ROC curves shown are for a single iteration of a 70–30 split of the data. (B) TF-binding prediction outperforms DNA sequence. Shown is a scatter plot comparing the AUCs achieved from TF-binding LR classifiers (y axis) and DNA sequence LR classifiers (x axis). Each point represents the mean of 10 computational experiments for one histone mark in one cell line. (C) H3K9ac profile at test set promoters. Shown on the Left is the mean H3K9ac profile at test set promoters predicted to be positive (green) and negative (red) for H3K9ac. Both predictions were performed on the same set of promoters. The dashed lines are predictions from sequence, and the solid lines are predictions from TFs. Notice the higher average of TF predicted positive marks. On the Right are heat maps of H3K9ac levels (white, low; red, high) at the individual promoters ordered by predicted positive probability (increasing along the y axis) as provided by sequence LR (Center) and by TF LR (Right).