Significance
The importance of viral icosahedral capsid-based nanoparticles (NPs) as cell-delivery vehicles is now being recognized. Virtually any specific protein and nucleic acid can be encapsidated together into a phage T4 capsid that can display surface-binding ligands for tissue targeting. In this study, T4 NPs packed in vivo with active cyclic recombination (Cre) recombinase and in vitro with fluorescent mCherry expression plasmid DNA were delivered into cancer cells. When released into cells together, the packaged active Cre recombinase within the capsid circularizes the packaged DNA of the linear expression plasmid, enhancing the expression of the mCherry gene. The efficient and specific packaging and unpackaging of DNA and active protein together into targeted cells has potential applications in targeted gene therapy and cancer therapy.
Keywords: terminase, DNA packaging, capsid decoration proteins, Soc, Hoc
Abstract
Packaging specific exogenous active proteins and DNAs together within a single viral-nanocontainer is challenging. The bacteriophage T4 capsid (100 × 70 nm) is well suited for this purpose, because it can hold a single long DNA or multiple short pieces of DNA up to 170 kb packed together with more than 1,000 protein molecules. Any linear DNA can be packaged in vitro into purified procapsids. The capsid-targeting sequence (CTS) directs virtually any protein into the procapsid. Procapsids are assembled with specific CTS-directed exogenous proteins that are encapsidated before the DNA. The capsid also can display on its surface high-affinity eukaryotic cell-binding peptides or proteins that are in fusion with small outer capsid and head outer capsid surface-decoration proteins that can be added in vivo or in vitro. In this study, we demonstrate that the site-specific recombinase cyclic recombination (Cre) targeted into the procapsid is enzymatically active within the procapsid and recircularizes linear plasmid DNA containing two terminal loxP recognition sites when packaged in vitro. mCherry expression driven by a cytomegalovirus promoter in the capsid containing Cre-circularized DNA is enhanced over linear DNA, as shown in recipient eukaryotic cells. The efficient and specific packaging into capsids and the unpackaging of both DNA and protein with release of the enzymatically altered protein–DNA complexes from the nanoparticles into cells have potential in numerous downstream drug and gene therapeutic applications.
Viral-based nanoparticles (NPs) that deliver cargo to specific targets continue to be of widespread interest for drug- and gene-delivery applications in human diseases and other areas of technology, such as diagnostic and cellular imaging (1–4). NPs derived from bacteriophage capsids are among the most attractive for these purposes (5–7). As NPs for medical applications, bacteriophage capsids have the advantages of (i) biocompatibility and demonstrated stability in the circulatory system and widespread delivery to tissues; (ii) absence of eukaryotic cell toxicity and of a preexisting immune response; (iii) ease and flexibility of molecular manipulation of the capsids and the encapsidated cargo; and (iv) easy accessibility of large amounts of material from fully characterized and nonpathogenic bacteria. Bacteriophage capsids and bacteriophages themselves vary tremendously in size and shape as well as complexity. The T4-like phages, which are among the largest and most complex of such bacteriophages, are particularly attractive because they are exceedingly well studied at the molecular and structural level. Thus, the large bacteriophage T4 capsid can serve in vaccine development, as displayed by surface decoration of full-length and active proteins (8–10), or for affinity studies using bipartite randomized peptide display libraries (Fig. 1) (11).
Fig. 1.
The T4 capsid-derived specific exogenous DNA plus protein packaging and eukaryotic cell delivery scheme. (A) DNA encoding a 10-amino acid N-terminal CTS peptide fused to the phage P1 Cre allows synthesis of CTS-Cre and targeting of the enzyme into the early core-scaffold of the T4 procapsid in vivo. Procapsid assembly and maturation-specific viral protease stabilize the procapsid, remove most of the scaffolding core as peptides, and remove the CTS peptide from Cre. Mutations in the viral terminase block DNA packaging and allow a mature but DNA-empty large Cre-containing procapsid to be highly purified from viral-infected bacteria. (B) In vitro packaging into the mature capsid of plasmid DNA containing mCherry driven by a CMV promoter and two loxP sites flanking an SfiI restriction enzyme site that allow the linearization required for packaging. The DNA is packaged into the procapsid by the ATP-driven terminase motor protein (gp17) with high efficiency. (C) The packaged Cre enzyme recircularizes the packaged linear plasmid DNA between the two loxP sites. The DNA-containing capsid is taken up by eukaryotic cells, here without displaying a specific peptide target, or into eukaryotic cells specifically using Soc and Hoc displayed peptides that have high affinity for the RP1 and RP2 receptors, respectively.
The phage T4 capsid has an additional unusual feature among bacteriophages: It is able to pack more than 1,000 dispersed and freely diffusing protein molecules together with the densely packed phage DNA (12, 13). Again, this protein-packaging feature has been thoroughly analyzed at the molecular level. Numerous exogenous proteins have been shown to be capable of encapsidation into the T4 procapsid precursor, and their copy numbers per particle have been determined (14). When fused with a short 10-amino acid N-terminal capsid-targeting sequence (CTS) peptide, the foreign protein is directed into the protein scaffolding-core precursor of the procapsid or prohead (12). Following procapsid maturation, and coupled to proteolytic processing by the highly specific encapsidated capsid maturation protease, in the absence of a mutated terminase packaging enzyme in vivo, the mature DNA-empty procapsid containing the encapsidated exogenous protein can be purified. The purified procapsid then can be packed with DNA by terminase in vitro. It has been shown that enzymes (e.g., micrococcal nuclease) and fluorescent proteins (e.g., GFP) retain and display activity within the empty procapsid and within the densely DNA-packaged capsid (15). About 200 copies of each of these proteins are packaged per particle, and DNA can be packaged in vitro in close proximity to the GFP within the purified GFP-containing procapsid (16, 17). The mature DNA-carrying capsid is taken up with high efficiency by eukaryotic cells and can be heavily labeled with fluorescent dye molecules (6). Additionally, the mature procapsid can bind high-affinity head outer capsid (Hoc) and small outer capsid (Soc) proteins that can be manipulated to display peptides or full-length proteins that confer the potential to enhance uptake into specific cell types (8–11).
In this study we show that the phage T4 capsid protein-packaging system allows post in vitro manipulation of DNA packaging by CTS-encapsidated cyclic recombination (Cre) recombinase. This prepackaged enzyme is able to recircularize within the capsid - the linear DNA that is packaged within the capsid and that is terminated by a loxP sequence at either end. Phage DNA packaging requires a linear DNA substrate, and Cre recircularization demonstrates that the packaged DNA can be manipulated further within the procapsid. Indeed recircularization enhances the biological activity of the encapsidated DNA in eukaryotic cells significantly, as measured by the expression of mCherry driven by a cytomegalovirus (CMV) promoter in plasmid mCherry (pmCherry).
Results
Packaging of Alexa 488-Labeled dsDNA into T4 Capsids.
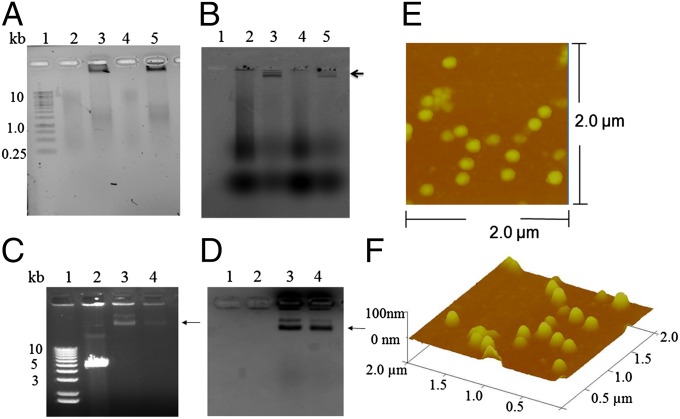
Previously, it was shown that the T4 gp17 large terminase subunit alone could package linear dsDNA of any sequence from 20–170 kb into purified procapsids with nearly 100% efficiency (Fig. S1A) but could not package dsRNA or RNA:DNA heteroduplexes (18) or circular DNA (19). To prepare heavily dye-labeled dsDNA for in vitro packaging and visualization of the encapsidated DNA by fluorescence microscopy, primer pairs for each Pseudomonas phage ф6 dsRNA genome segment (L, M, and S) were used to synthesize each cDNA strand separately by means of reverse transcriptase. This synthesis was followed by RNase treatment, heating, and finally annealing of the complementary cDNA strands by decreasing the temperature incrementally to 4 °C. For the reverse transcription, a 2:1 mixture of modified dCTP [5-aminohexylacrylamido–dCTP; (aha)-dCTP] and unmodified dCTP was incorporated into the DNA fragments. The modified base allows reaction with N-hydroxysuccinimide (NHS) ester dyes. In theory 30% of genomic sequences should contain aha-dCTP based on a 45% adenine (A) and thymine (T) nucleotide base content obtained from the actual bases within the targeted ф6 genomic sequences. After annealing, the resulting aha-dCTP–modified dsDNA was purified and mixed with Alexa 488-NHS ester at room temperature for 1 h. Approximately 5–10% of the bases were labeled with Alexa 488 dyes incorporated into the DNA fragments based on the absorbance at 495 nm and on the DNA concentration estimated by absorption at 260 nm. Electrophoresis results also show that reverse-transcribed ssDNA containing aha-dCTP and labeled dyes results in a mixed population with dsDNA lengths of 0.5–10 kb after annealing, possibly as a result of the aha-dCTP modification. Although a single expected distinct dsDNA target size (3.3 kb for L-segment dsDNA and 1.5 kb for S-segment dsDNA) is not seen, the majority of the L-segment and S-segment dsDNA population does localize within 1–5 kb (Fig. 2A). A significant portion of this heterogeneous DNA can be packaged into procapsids in vitro, as seen in the low-mobility band retained in front of the loading well indicating that this dsDNA is packaged into the procapsids and is resistant to DNase treatment (Fig. 2 A and B). Alexa 488-labeled M-segment dsDNA also was prepared and packaged into the procapsids for subsequent cellular uptake experiments. The high-mobility dye front appearing in Fig. 2B is free Alexa 488 that remained even after membrane filtration. The residual free Alexa 488 dyes with net negative charges are not cell permeable and washed off after the treatment. They had no effect on the efficiency of cellular uptake (6). Here, we demonstrate for the first time (to our knowledge) that the bulky DNA-containing, Alexa 488–aha-dCTP–modified base can be packaged, although with reduced efficiency, by the T4 terminase in vitro. Furthermore, the integrity of the procapsids containing the packaged linear mCherry DNA and dye-labeled DNA was demonstrated by the sharp protein bands, which are at the same positions as the shifted DNA bands (Fig. 2 C and D). Atomic force microscopy (AFM) also confirmed that the purified procapsids were intact and stable (Fig. 2 E and F). To carry out in vitro DNA packaging, 2 µg of linear plasmid DNA or dye-labeled DNA and ∼1.6E+10 capsids were used in a 20-µL reaction. Our result indicated that 2 µg of linear plasmid DNA were completely packaged into 1.6E+10 procapsids (Fig. S1A), so each capsid contained ∼24–75 copies of the packaged DNA, depending on the size of DNA (Table S1); packaging of multiple pieces of DNA per procapsid has been demonstrated previously (17). However, packaging of dye-labeled dsDNA was less efficient (∼50%) (Fig. 2A). This estimate of efficiency was used to determine the numbers of DNA per capsid.
Fig. 2.
Packaging of fluorescent dsDNAs into T4 procapsids and assessment of the integrity of T4 NPs. (A) Ethidium bromide staining viewed on a biospectrum imaging system (UVP). The gel was run for only a short time and adjusted to a bright background with less contrast to visualize the low molecular weight smear DNA. (B) The unstained gel viewed on a Typhoon imager using an Alexa 488 dye filter. The L-segment (3,320 bp) of ф6 dsRNA was reverse transcribed using aha-dCTP followed by conjugation with Alexa 488 NHS ester (Amersham), as shown in lane 2, and the dye-labeled L-segment dsDNA was packaged into control T4 procapsids lacking the Cre enzyme, as shown in lane 3 (arrow near the loading well). The S-segment (1,520 bp) of ф6 dsRNA (lane 4) was reverse transcribed by the same procedure. The dye-labeled S-segment dsDNA is shown in lane 5. Lane 1 shows the 1-kb DNA size marker (Thermo Scientific). The arrow indicates the position of packaged DNA retained by procapsids near the top of the agarose gel. (C and D) Images of the same agarose gel viewed on a Gel Doc EZ imaging system (Bio-Rad). (C) Ethidium bromide staining image with more contrast (dark background). The agarose gel was run for a longer time to allow the packaged DNA to migrate further for visualizing the packaged DNA on the top (indicated by the arrow) due to slow migration of the large procapsids. The doublet bands possibly result from two different charges of the procapsids. The unpackaged linear EGFP DNA with a size of ∼5.5 kb is shown in lane 2, and packaged EGFP DNA with a size >10 kb is shown in lane 3. Packaged Alexa 480-labeled L-segment dsDNA with a size >10 kb is shown in lane 4. (D) Coomassie blue staining image of C. T4 capsids in lanes 3 and 4 were stained with Coomassie blue as indicated by the arrow. (E and F) Purified T4 proheads for DNA packaging were visualized by AFM with a scanning area of 2.0 × 2.0 µm. A 3D view is shown in F.
Cellular Uptake of T4 Capsids Packaged with the Fluorescent dsDNA.
The procapsids loaded with packaged fluorescent L-, M-, and S-segment dsDNA fragments then were subjected to cellular uptake experiments using lung cancer epithelial A549 cells. Cells containing internalized fluorescent procapsids become fluorescent and show different histogram patterns resulting from varied fluorescent cell populations separated by a flow cytometer as described in SI Materials and Methods. The fluorescent cell population is concentration-dependent, because the cells treated with higher amounts of T4 procapsids packaged with Alexa 488-labeled L and S dsDNA segments (LL- and SL-DNA, respectively) appear to have more fluorescent cells, shifting the histogram curve toward the right (Fig. 3 A and B). The internalized Alexa 488-labeled M (ML-) dsDNA fragments (∼2.0 kb) packaged within the procapsids were subjected to uptake experiments for 6 h, followed by nucleus staining and epifluorescent microscopy. The treated cells were washed extensively with PBS to remove residual free dyes present in the reaction. The green spots representing the packaged Alexa 488-labeled M-dsDNA are visualized within the cells relative to nuclei (Fig. 3 C–E). Interestingly, multiple small, green fragments (small arrows in Fig. 3C) were observed also, suggesting that the packaged M-dsDNA fragments were released from the procapsids inside A549 cells. The treatment of cells with the dye-stained DNA-containing procapsids is not toxic to cells, which remain intact, as shown in Fig. 3F. Cellular uptake of T4 NPs in different cell types was observed previously (6). The endocytosis pathways may be different in different cell types, and our preliminary data from inhibitor studies (Fig. S2) indeed suggest that uptake of T4 NPs into A549 cells occurs through multiple pathways.
Fig. 3.
Assessment of the cellular uptake of T4 procapsids containing packaged Alexa 488-labeled L-segment– and S-segment–derived ф6 dsDNA. (A and B) Histograms of the flow cytometry with varied concentrations of T4 procapsid-packaged Alexa 488-labeled L-segment (LL) and S-segment (SL) dsDNA, respectively. (C) The internalized Alexa 488-labeled M-segment (ML) dsDNA (∼2,000 bp, indicated by small arrows) within A549 cells was visualized by epifluorescent microscopy. (D) DAPI staining for nuclei (white arrow). (E) Overlay of C and D. (F) Bright-field image showing whole cells with nuclei indicated by the black arrow. (Scale bars, 10 µm.)
Transformation of Packaged Linear mCherry DNAs Isolated from T4 Procapsids.
To measure the transformation efficiency of the packaged linear plasmid DNA, the packaged DNA was phenol extracted and transformed into Escherichia coli. Plasmids, such as pmCherry-C1, carry the fluorescent protein gene and are used for packaging and conducting transformation. mCherry-C1 plasmids contain two origins of replication (ori), one for E. coli and one for eukaryotic cells, to permit replication in both types of cells; in addition, the plasmids contain two selectable antibiotic resistance markers for selecting the E. coli or eukaryotic recipients. The plasmids also contain an upstream CMV promoter followed by the mCherry gene for expression of fluorescent mCherry in eukaryotic recipients only. To prepare linear mCherry-C1 for packaging assays, circular plasmids were cut with SfiI restriction enzyme without disrupting the ori genes, antibiotic marker genes, or CMV promoter. Initially, the efficiency of E. coli transformation from packaged linear mCherry-C1 isolated from wild-type (Cre−) T4 procapsids was assessed using a chemical transformation protocol and appeared to be less than 102 cfu/µg because of nonreplicable linear dsDNA. To improve the transformation efficiency of packaged linear mCherry-C1 DNA, Cre recombinase was prepackaged into the procapsids by fusion to the CTS (Fig. 4A). Our results indicated that DNase incubation treatment has no effect on the amount of prepackaged Cre recombinase retained per purified procapsid (Fig. 4A). Two specific Cre-binding sequences (loxP) were inserted around an SfiI site to create pmCherryC1–loxP (Fig. 4B). Linear mCherryC1–loxP dsDNA generated by SfiI digestion and T4 procapsids containing Cre+ then were used for DNA-packaging assays. The packaged linear loxP DNA was circularized inside the Cre+ procapsids, and the resulting circular DNA migrated much more slowly than the packaged linear loxP DNA within Cre− procapsids (Fig. 4C). Consistently, the phenol-extracted packaged mCherryC1–loxP DNA from Cre+ procapsids shows the appearance of the circular form of mCherryC1–loxP and the disappearance of linear dsDNA (Fig. 4D), whereas no circular form appears in Cre− procapsids. The isolated plasmid DNAs were used to transform recombination deficient E. coli chemically by heat shock. The circular pmCherryC1–loxP from Cre+ procapsids generated many colonies, whereas, as expected, the linear plasmid DNA from Cre− procapsids generated none. In addition the transformation efficiency (cfu per milligram) using XL1 Blue cells and electroporation is 40-fold higher for the Cre+ procapsid-packaged DNA (Fig. 4D, lane 5) than for the linear DNA isolated from Cre− procapsids (Fig. 4 D, lane 4, and E). Overall, our results strongly suggest that the encapsidated Cre recombinase is active and recircularizes the linear pmCherryC1–loxP DNA within the capsid by recombination between the two loxP DNA sequences at the ends of the linearized DNA.
Fig. 4.
The recircularization of packaged linear mCherryC1–loxP plasmid DNA within procapsids containing Cre recombinase. (A) An immunoblot using anti-Cre antibody (Novagen) shows that the CTS–Cre recombinase is packaged into Cre+ T4 procapsids (lanes 2 and 3) but is not detected in a comparable amount of Cre− T4 procapsids (lane 1). As shown in lane 3, DNase treatment did not affect the Cre+ content internalized within the procapsids. Lane 4 is a molecular size marker, and lanes 5–7 show increasing amounts of purified Cre recombinase (Novagen) as quantification standards and positive controls. (B) The mCherryC1–loxP plasmid was obtained by inserting two loxP sites linked by an SfiI restriction enzyme site (as indicated in the sequence box) into a commercial mCherry-C1 plasmid (Clontech). (C) Phenol-extracted packaged mCherry–loxP DNA within Cre− and Cre+ procapsids. Lane 2 shows unpackaged SfiI-linearized pmCherry–loxP DNA. Lane 3 shows the slower-moving circular form of packaged pmCherryC1–loxP DNA isolated from Cre+ procapsids. The circular mCherry–loxP DNA (arrow) migrated more slowly than the packaged linear pmCherryC1–loxP DNA isolated from Cre− procapsids in lane 4. (D) Packaged SfiI-linearized plasmid DNA from Cre− and Cre+ T4 procapsids. Purified pmCherryC1–loxP DNA and phenol-extracted packaged DNA isolated from Cre− and Cre+ T4 procapsids were assessed for E. coli transformation and compared by agarose electrophoresis. Lanes 2 and 3 show unpackaged linear pmCherryC1–loxP DNA cut by SfiI moving faster than the uncut circular pmCherryC1–loxP. Packaged linear pmCherryC1–loxP DNA remained linear in Cre− T4 procapsids (lane 4), whereas the packaged linear pmCherryC1 DNA was circularized in Cre+ T4 procapsids (arrows in lane 3 in C and lane 5 in D). Lane 1 is the 1-kb+ DNA size marker (Life Technologies, Inc.). (E) An ∼40-fold increase in transformation efficiency was exhibited by the recircularized pmCherryC1–loxP DNA isolated from Cre+ T4 procapsids relative to Cre− T4-packaged DNA (D, lane 4 vs. lane 5). Error bars represents SE; n = 2; *P < 0.05, t test.
Expression of Fluorescent mCherry Protein from Packaged mCherryC1–loxP DNA.
To investigate the release and expression of DNA internalized within procapsids into A549 cells, linearized plasmid DNA, mCherryC1–loxP, was used for an in vitro DNA-packaging assay, followed by treatment of A549 cells with Cre− and Cre+ procapsids containing the packaged linear plasmids. A549 cells were treated with T4 procapsids loaded with the linear DNA for 6 h, followed by 24-h incubation. Fluorescent mCherry protein from released linear plasmids inside A549 cells was measured using flow cytometry. Previously, we demonstrated that the uptake of Alexa 546-conjugated T4 procapsids by A549 cells made the cells fluorescent and distinguishable from nonfluorescent cells (6). The fluorescent cells shift the histogram of the cell population toward the right in a concentration-dependent manner (Fig. 5A). Consistent with the transformation results, large, fluorescent A549 cells resulting from internalized circular mCherryC1–loxP in Cre+ procapsids shift the histogram toward the right (Fig. 5 B and C), suggesting that the packaged DNA is released and expressed in the recipients (Fig. 5 B and C). There is no shift in A549 cells containing internalized linear mCherryC1–loxP in Cre− procapsids (Fig. 5D). The median and mean fluorescent intensities were significantly higher in A549 cells treated with the Cre+ procapsids containing mCherryC1–loxP DNA (Fig. S3 and Table S2). In addition to mCherryC1–loxP, linear mCherry-C1 and EGFP-C1 DNA also were used for the DNA-packaging and gene-expression measurements. However, despite active CMV promoters, the expression of fluorescent protein is insignificant; there is no significant shift of fluorescent cells population in FACS histogram relative to the control, and there is very weak fluorescent protein expression in A549 cells resulting from nonreplicable linear plasmid DNA (Fig. S1 B and C and Table S3). If the targeting ligand were displayed on the surface of procapsids to enhance the cellular uptake, the expression of linear plasmids could be improved significantly (10).
Fig. 5.
Flow cytometry measurement of fluorescent protein expression in A549 cells from T4-packaged plasmid mCherryC1–loxP DNA. (A) A fluorescent control shows that treatment of the Alexa 546-labeled T4 NPs (6), which were obtained by conjugating the dyes to the capsid proteins, produced a concentration-dependent shift in the fluorescent population. The rightward shift of the curves (red and blue traces) indicates a higher cell fluorescent intensity with a larger portion of fluorescent cells. SFM without the addition of any T4 NPs (black trace) served as a negative control. (B and C) A larger fluorescent cell population was observed in A549 cells treated with the 4.0E+9 Cre+ T4 procapsids (red trace in B) and 8.0E+9 T4 procapsids containing circular pmCherryC1–loxP (red trace in C) relative to SFM (black traces in B and C), indicating the expression of fluorescent mCherry protein in A549 cells from the packaged circular pmCherryC1–loxP DNA. (D) A549 cells treated with the linear DNA packaged in Cre− T4 NPs (red and blue traces) showed no difference in cell fluorescent intensity and population (i.e., no right shift) relative to cells treated with SFM (black trace).
Assessment of Cell Viability of Cre Procapsid-Treated A549 Cells and Cytotoxicity of Cre+ Procapsids for A549 Cells.
Previously, we demonstrated that T4 procapsids are highly biocompatible with eukaryotic cells, and the uptake of dye-labeled Cre− procapsids does not result in any cytotoxicity (6). Moreover, we measured cell viability and proliferation using the tetrazolium compound 3-[[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide] (MTT) 24 h after cell uptake of Cre− procapsids. Either dye-labeled or unlabeled Cre− T4 NPs were used for the cell uptake. Cells treated with either unlabeled or dye-labeled T4 procapsids showed 100% viability after normalizing to the untreated cells (Fig. 6A). Consistently, the cells treated with the Cre− procapsids appeared adherent and attached to each other without dead cells floating in the medium (Fig. 6B), comparable to the untreated control (Fig. 6C). In contrast, the uptake of Cre+ procapsids caused extensive cell death starting at 24 h. Dead cells were visualized as detached, opaque, and smaller and rounder vesicle-like bodies floating in the medium. Only a small portion of cells were still attached and appeared intact (Fig. 6D). The lethal effect of prepackaged Cre recombinase is good evidence that the encapsidated Cre recombinases had been released into the cytoplasm of the recipient cells (20).
Fig. 6.
Assessment of viability of A549 cells treated with Cre− procapsids and of the cytotoxicity induced by the treatment of Cre− T4 NPs. (A) The cell uptake of unlabeled Cre− or Alexa 647-labeled Cre− (A647-T4) procapsids showed no cytotoxicity for A549 cells, which appeared 100% viable. (B–D) There is no apparent cytotoxicity in A549 cells treated with Cre− T4 procapsids (B) relative to the untreated cells (C), whereas most of the cells appeared dead (i.e., as rounded opaque spots) after treatment with Cre+ T4 procapsids (D). All bright-field images of A549 cells were obtained using a 20× objective. (Scale bars, 50 µm.)
Discussion
T4 capsids display a large surface area and have a large cargo capacity for carrying protein and DNA molecules for cell delivery. Previously, we demonstrated that more than 104 dye molecules per capsid were chemically conjugated to both the inner and outer surfaces of the T4 capsid, yielding highly fluorescent T4 NPs that were used for imaging and flow cytometry applications after internalization by cancer cells (2, 6). In this study, we further demonstrated the imaging of highly fluorescent T4 capsids that result from encapsidating multiple modified bases conjugated to reactive dyes in dsDNA synthesized by reverse transcriptase. Also, for the first time (to our knowledge), we have shown that the heavily dye-modified base containing dsDNA can be packaged into procapsids with relatively high efficiency by the viral packaging terminase in vitro. We also demonstrated that the fluorescent dsDNA is released into the cell cytoplasm after internalization within cancer cells, as indicated by the length of the imaged fluorescent DNA fragments (∼1 µm; arrows in Fig. 3C), which was 10 times longer than the long axis of the capsid (0.1 µm). Our results are consistent with a recent study that shows that T4 capsids displaying cell receptor-interacting proteins can be internalized into target cells effectively and can deliver linear GFP dsDNA for protein expression inside the target cells (10). Although T4 capsids that do not display the cell receptor for targeting cells also deliver the linear DNA, the efficiency is not sufficient to show the histogram shift measured by flow cytometry. However, we did see higher median and mean values in Cre− T4–treated samples than in a negative control. [Serum-free media (SFM) are listed in Table S1.]
With a few exceptions, such as Cowpea Chlorotic Mottle Virus (CCMV) (21) and phage T4 (13), the ability to package protein inside capsids usually is limited in most viral-based delivery systems. CCMV can package the protein by in vitro assembly controlled by pH, whereas T4 has a well-characterized CTS that allows numerous proteins to be preassembled in vivo into the DNA-free precursor procapsid at a high copy number (12–14). Using this established system, we further demonstrated that the T4 capsid NP gene-expression system delivers both specific proteins and DNAs simultaneously into targeted recipient eukaryotic cells. In contrast to protein delivery through capsid surface display (10), the protein is sequestered inside the capsid for cell delivery in our T4 capsid NP delivery system. Our system has the advantage of being able to package both active enzymes and cognate DNAs inside the capsids, delivering both together into targeted recipient cells. Specifically, we have shown CTS-encapsidated Cre recombinase has been preassembled into procapsids, and procapsids containing Cre recombinase can be purified and then packaged in vitro with linear DNA containing two loxP recombination target sites for Cre. The encapsidated Cre enzyme is active within the procapsid and recircularizes the packaged double loxP site containing linear DNA within the procapsid by recombination between the two loxP site sequences. This recombination is shown by gel electrophoresis of the procapsid-extracted DNA and by enhanced transformation of bacteria using the circularized packaged procapsid DNA (Fig. 4 C and D). In addition the expression of an mCherry fluorescent protein gene from a CMV promoter is greatly enhanced after the uptake and release of the circular restructured DNA into eukaryotic cells. The toxicity of the Cre protein in the recipient cancer cells indicates that both the Cre protein and the restructured loxP circular DNA are transferred into cancer cells. In combination with the targeted delivery strategy, the high copy number of Cre+-containing T4 NPs also may be used as therapeutic NPs to kill cancer cells specifically. Potentially, this Cre protein–loxP DNA combined NP also can deliver DNA into preexisting specific loxP sites in cells or loxP-like chromosomal sites in specific targeted cells (20).
Although the Cre recombinase released from internalized Cre+ procapsids causes extensive cell death late after delivery (Fig. 6D), it should be possible to assess the ability of the Cre+ loxP T4 NPs to integrate recircularized DNAs containing the double loxP site into viable cells with chromosomal loxP sites by optimizing conditions (e.g., a lower infection ratio of T4 capsids to cells, shorter incubation time, or less Cre enzyme per capsid to reduce the cell death in the recipient eukaryotic cells). In fact, the amount of packaged protein per particle already has been regulated using E. coli suppressor mutants that translate UAG-containing CTS transcripts with low or high efficiency (15). In these experiments we measured ≥50 copies per procapsid (Fig. 4A), consistent with previous studies (14, 15). With additional features that are within the reach of current technologies, procapsids containing multiple copies of the Cre enzyme and up to 170 kb of linear DNA of any sequence packaged in vitro to fill the procapsid can be used for cell targeting. Potentially, the NPs (i) can target specific eukaryotic cell receptors by targeting peptides or full-length proteins displayed from both the 155 copies of Hoc protein per capsid and the 810 copies of Soc protein per capsid (9, 10); (ii) can deliver the recircularized Cre DNAs with loxP at either end into the nucleus together with the Cre enzyme itself for superior gene expression; and (iii) can combine delivery of the Cre enzyme with delivery of the loxP sequence-specific DNA to target specific chromosomal sites for gene expression and potentially for targeted gene repair. Because there are natural Cre-targeting sites in unmodified genomes, it is possible that advances in Cre recombinase-related design might allow quite specific targeting to unmodified inactive or damaged DNA in chromosomes (20).
By taking advantage of the ability to encapsidate both specific proteins and DNAs, many additional potential applications of this T4 capsid NP targeting and delivery system can be explored. One specific example follows upon our demonstration that comparable CTS-expression phages target ∼50 copies of λ exonuclease into a procapsid or mature phage capsid. The procapsids are excellent receptacles for in vitro DNA packaging, and the λ exonuclease also is active within proheads on linear plasmid DNA that is packaged in vitro. By also adding λβ protein to the proheads, as can be done readily using the CTS approach that can target two proteins together into a single procapsid (15), we can determine whether the essential features of the λ-red recombineering system (λ exonuclease protein plus λβ protein activities) work when delivered into eukaryotic cells, as they do in bacterial cells (22), and as do the Cre recombinase and multiple other phage recombination enzymes (20). Limited digestion at both 5′ ends of the packaged linear DNA by the packaged λ exonuclease should allow us to test for full DNA sequence-based homologous recombination by the λ-red system in both prokaryotic and eukaryotic cells. This approach could allow a full DNA sequence- and protein enzyme-based homologous recombineering approach to replacement gene therapy by targeting any DNA sequence gene in the eukaryotic chromosome. Overall, the T4 capsid NP gene expression and protein delivery system may be complementary to or used in conjunction with RNA Cas and taran nuclease-based gene therapy approaches (23).
Materials and Methods
Enzymes and Reagents.
All restriction endonuclease enzymes were purchased from New England Biolabs, and chemical reagents were obtained from Sigma-Aldrich unless specified in the text. DNase, RNase, and DNA purification kits were acquired from Qiagen, Inc. Bacterial medium and cell-culture medium were purchased from Life Technologies.
Preparation of the Cre+ and Cre− Procapsids.
DNA encoding the cre gene was synthesized by PCR using a phage λi434cre+ DNA template using PCR primers flanked with HindIII and EcoRI sequences, respectively. The cloning of the cre gene into PJM252 and recombinant phage selection for preparation of Cre+ and Cre− procapsids are described in detail in SI Materials and Methods.
Preparation of T4 Procapsids AFM.
Purified T4 procapsids were resuspended in 10 mM Tris⋅HCl with 2 mM MgCl2 at pH 7.5 at a concentration of 2.0E+10 procapsids/mL Five microliters of the prepared T4 NPs were deposited on freshly cleaved mica and kept in a humid environment at room temperature for 12 h. After the T4 NPs were adsorbed onto the surface, the substrates were allowed to dry at room temperature and then were rinsed carefully with Milli-Q water. AFM imaging and data analysis were performed according to a previously established protocol (24). Details are given in SI Materials and Methods.
Preparation of Dye-Labeled dsDNA.
Three Pseudomonas phage ф6 dsRNA genome fragments, L, M, and S (NC003714, NC003715, and NC003716) were isolated and used for reverse transcription in the presence of the designed primers specific for each fragment, followed by labeling with Alexa 488. See SI Materials and Methods for detailed information.
Preparation of pmCherryC1–loxP DNA for in Vitro DNA Packaging.
Fluorescent mCherry protein expression plasmid, pmCherry-C1, was obtained from Clontech Lab. The plasmid was linearized using SfiI without interrupting the CMV promoter for RNA transcription. Primers containing the loxP sequence, 5′−ATAACTTCGTATAGCATAC-ATTATACGAAGTTAT-3′, and the flanking SfiI sites were used to amplify the whole plasmid. The amplified plasmid DNA then was cut with SfiI and recircularized by self-ligation. The resulting plasmids were sequenced to verify the loxP sequences. Linearized pmCherryC1–loxP DNA was used for packaging assays.
DNA Packaging Assays.
Approximately 1.6 × 1010 (determined by OD280nm) purified procapsids were incubated with purified terminase and linear dsDNA (1–3 µg) in the DNA packaging reaction buffer at room temperature for 1 h as described by Dixit et al. (25, 26).
Transformation of Phenol-Extracted T4-Packaged DNA.
To confirm the circularization of packaged linear loxP DNA, DNA packaged into both wild-type Cre− procapsids and procapsids derived from 9 aminoacridine were treated with DNase to remove unpackaged DNA, were incubated with proteinase K:EDTA:SDS to release packaged DNA as described by Black and Peng (19), and then were extracted by phenol chloroform. The purified DNA was resuspended in 5 µL of water for electroporation (XL1 Blue) or chemical transformation (DH10β). The efficiency of electroporation transformation was calculated as cfu per microgram.
Uptake of T4 NPs by A549 Cells.
T4 NPs packaged with either dye-labeled DNA or with linear pmCherry-C1 and pmCherryC1–loxP DNA were added to A549 cells in an estimated ratio of 50,000 particles per cell for 15–18 h. Cells treated with fluorescent T4 NPs then were fixed with 4% (vol/vol) formaldehyde for 15 min, followed by staining with 5 µg/mL of DAPI at room temperature for 15 min and washing five times with PBS. Cells treated with T4 NPs containing linearized DNA were incubated further in growth medium for 48 h after washing. Cells then were fixed and DAPI stained as previously described. The images were obtained using an epifluorescent microscope (Nikon TE2000) at a magnification of 60× and were analyzed using the Nikon Imaging software, NIS-Elements AR 3.22.
Cell Viability and Proliferation Measurement for Dye-T4–Treated Cells.
Cells were treated with dye-T4/T4 NPs at various concentrations ranging from 1.0E+4–1.0E+6 for 4 h, followed by washing three times with PBS and the addition of new growth medium. The cells were grown for another 24 h, and the tetrazolium compound MTT was added for 1 h. Cell concentrations were measured by the absorbance at 620 nm. Untreated cells served as negative control. Triplicate measurements were used for each treatment and for the negative control.
Flow Cytometry for Sorting and Quantification of the Fluorescence of A549 Cells.
A549 lung cancer cells with 80% confluence on 24-well tissue-culture plates were treated with 2 × 1010 T4 NPs and T4 NP derivatives for 6 h. After incubation, the cells were washed twice with 1× PBS and harvested with trypsin-EDTA. The cells then were washed five times with 1× PBS, resuspended in 1× PBS, and run on an Accuri C6 flow cytometer using standard lasers and filters for GFP (FL-1), and mCherry (FL-2). For each sample 2 × 104 events were collected in a gate corresponding to the cell population. To determine the percentage of live cells, a scatter plot showing the intensity of the live/dead reagent (FL-4) vs. forward scatter (FSC) was gated for the population with the lowest fluorescence (live cells). The percentage of cells positive for the fluorescent T4 NPs was calculated using the histogram subtraction tool on FSC Express V3 (De Novo Software).
Supplementary Material
Acknowledgments
We thank the late Dr. N. Sternberg for his gift of phage λi434cre+, Dr. Leonard Mindich for his gift of phage ф6, and Drs. J. A. Thomas and J. M. Mullaney for their valuable comments on the manuscript. J.L.L. and K.L.R. were supported by Naval Research Laboratory (NRL) funding, E.Q. was supported by the NRL Science and Engineering Apprenticeship Program, and L.W.B. and A.B.D. were supported by National Institutes of Health Grant RO1 AI011676.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321940111/-/DCSupplemental.
References
- 1.Manchester M, Singh P. Virus-based nanoparticles (VNPs): Platform technologies for diagnostic imaging. Adv Drug Deliv Rev. 2006;58(14):1505–1522. doi: 10.1016/j.addr.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 2.Robertson KL, Liu JL. Engineered viral nanoparticles for flow cytometry and fluorescence microscopy applications. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2012;4(5):511–524. doi: 10.1002/wnan.1177. [DOI] [PubMed] [Google Scholar]
- 3.Manchester M, Steinmetz NF. Viruses and nanotechnology. Preface. Curr Top Microbiol Immunol. 2009;327:v–vi. [PubMed] [Google Scholar]
- 4.Wu Z, et al. Development of viral nanoparticles for efficient intracellular delivery. Nanoscale. 2012;4(11):3567–3576. doi: 10.1039/c2nr30366c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashley CE, et al. Cell-specific delivery of diverse cargos by bacteriophage MS2 virus-like particles. ACS Nano. 2011;5(7):5729–5745. doi: 10.1021/nn201397z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robertson KL, Soto CM, Archer MJ, Odoemene O, Liu JL. Engineered T4 viral nanoparticles for cellular imaging and flow cytometry. Bioconjug Chem. 2011;22(4):595–604. doi: 10.1021/bc100365j. [DOI] [PubMed] [Google Scholar]
- 7.Li K, et al. Chemical modification of M13 bacteriophage and its application in cancer cell imaging. Bioconjug Chem. 2010;21(7):1369–1377. doi: 10.1021/bc900405q. [DOI] [PubMed] [Google Scholar]
- 8.Ren ZJ, et al. Phage display of intact domains at high copy number: A system based on SOC, the small outer capsid protein of bacteriophage T4. Protein Sci. 1996;5(9):1833–1843. doi: 10.1002/pro.5560050909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Black LW, Rao VB. Structure, assembly, and DNA packaging of the bacteriophage T4 head. Adv Virus Res. 2012;82:119–153. doi: 10.1016/B978-0-12-394621-8.00018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tao P, et al. In vitro and in vivo delivery of genes and proteins using the bacteriophage T4 DNA packaging machine. Proc Natl Acad Sci USA. 2013;110(15):5846–5851. doi: 10.1073/pnas.1300867110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malys N, Chang DY, Baumann RG, Xie D, Black LW. A bipartite bacteriophage T4 SOC and HOC randomized peptide display library: Detection and analysis of phage T4 terminase (gp17) and late sigma factor (gp55) interaction. J Mol Biol. 2002;319(2):289–304. doi: 10.1016/S0022-2836(02)00298-X. [DOI] [PubMed] [Google Scholar]
- 12.Mullaney JM, Black LW. Capsid targeting sequence targets foreign proteins into bacteriophage T4 and permits proteolytic processing. J Mol Biol. 1996;261(3):372–385. doi: 10.1006/jmbi.1996.0470. [DOI] [PubMed] [Google Scholar]
- 13.Hong YR, Black LW. Protein folding studies in vivo with a bacteriophage T4 expression-packaging-processing vector that delivers encapsidated fusion proteins into bacteria. Virology. 1993;194(2):481–490. doi: 10.1006/viro.1993.1287. [DOI] [PubMed] [Google Scholar]
- 14.Mullaney JM, Black LW. Bacteriophage T4 capsid packaging and unpackaging of DNA and proteins. Methods Mol Biol. 2014;1108:69–85. doi: 10.1007/978-1-62703-751-8_5. [DOI] [PubMed] [Google Scholar]
- 15.Mullaney JM, Black LW. Activity of foreign proteins targeted within the bacteriophage T4 head and prohead: Implications for packaged DNA structure. J Mol Biol. 1998;283(5):913–929. doi: 10.1006/jmbi.1998.2126. [DOI] [PubMed] [Google Scholar]
- 16.Mullaney JM, Thompson RB, Gryczynski Z, Black LW. Green fluorescent protein as a probe of rotational mobility within bacteriophage T4. J Virol Methods. 2000;88(1):35–40. doi: 10.1016/s0166-0934(00)00166-x. [DOI] [PubMed] [Google Scholar]
- 17.Sabanayagam CR, Oram M, Lakowicz JR, Black LW. Viral DNA packaging studied by fluorescence correlation spectroscopy. Biophys J. 2007;93(4):L17–L19. doi: 10.1529/biophysj.107.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oram M, Sabanayagam C, Black LW. Modulation of the packaging reaction of bacteriophage t4 terminase by DNA structure. J Mol Biol. 2008;381(1):61–72. doi: 10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Black LW, Peng G. Mechanistic coupling of bacteriophage T4 DNA packaging to components of the replication-dependent late transcription machinery. J Biol Chem. 2006;281(35):25635–25643. doi: 10.1074/jbc.M602093200. [DOI] [PubMed] [Google Scholar]
- 20.Turan S, Bode J. Site-specific recombinases: From tag-and-target- to tag-and-exchange-based genomic modifications. FASEB J. 2011;25(12):4088–4107. doi: 10.1096/fj.11-186940. [DOI] [PubMed] [Google Scholar]
- 21.Douglas T, Young M. Host-guest encapsulation of materials by assembled virus protein cages. Nature. 1998;393(6681):152–155. [Google Scholar]
- 22.Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: A homologous recombination-based method of genetic engineering. Nat Protoc. 2009;4(2):206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li JF, et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31(8):688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Archer MJ, Liu JL. Bacteriophage t4 nanoparticles as materials in sensor applications: Variables that influence their organization and assembly on surfaces. Sensors (Basel) 2009;9(8):6298–6311. doi: 10.3390/s90806298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dixit AB, Ray K, Black LW. Compression of the DNA substrate by a viral packaging motor is supported by removal of intercalating dye during translocation. Proc Natl Acad Sci USA. 2012;109(50):20419–20424. doi: 10.1073/pnas.1214318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dixit A, Ray K, Lakowicz JR, Black LW. Dynamics of the T4 bacteriophage DNA packasome motor: Endonuclease VII resolvase release of arrested Y-DNA substrates. J Biol Chem. 2011;286(21):18878–18889. doi: 10.1074/jbc.M111.222828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.