Fig. 4.
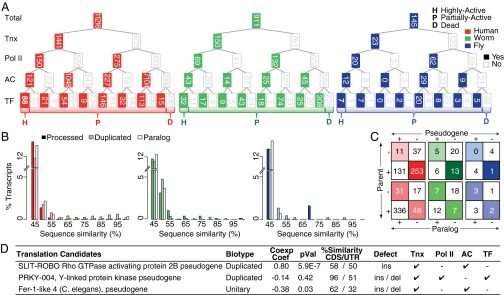
Pseudogene activity. (A) Distribution of pseudogenes as a function of various activity features: transcription (Tnx), active chromatin (AC), and presence of active Pol II and TF binding sites in the upstream region. (B) Conservation of the upstream sequences in processed and duplicated pseudogenes compared with paralogs. (C) Conservation of an upstream sequence activity mark (H3K27Ac) in pseudogene-parent pairs vs. parent-paralogs. +, active H3K27Ac; −, inactivity. We find that the majority of parent–paralog pairs have coordinated H3K27Ac activity (larger diagonal values) as opposed to parent–pseudogene pairs (larger off-diagonal values). (D) Functional pseudogene candidates with translation evidence.
