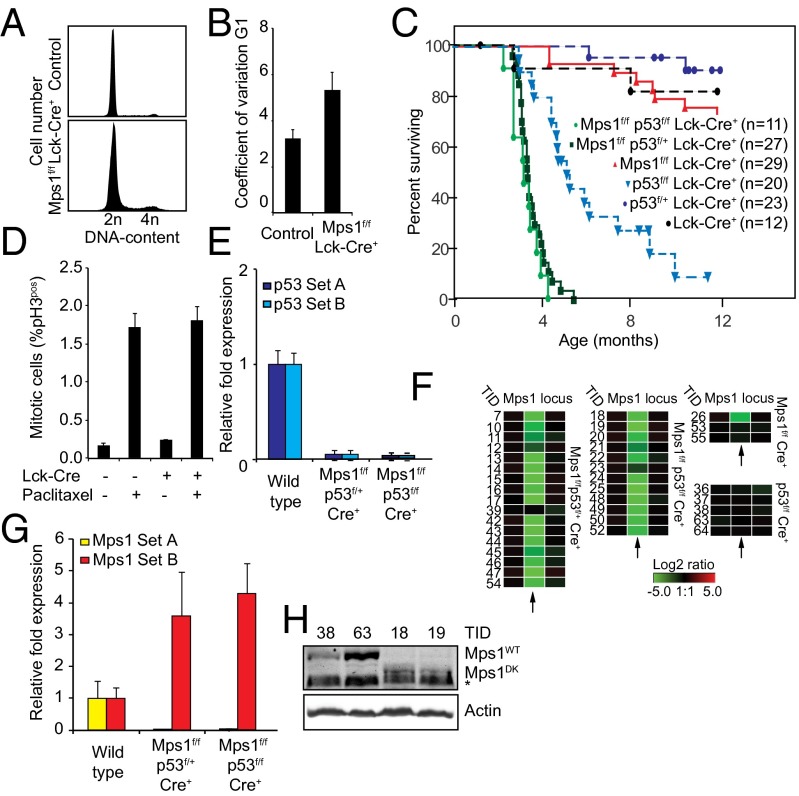
Fig. 2.
Mps1 truncation provokes aneuploidy in vivo and decreases T-ALL latency in a p53-compromised background. (A) Distribution of DNA content in control and Mps1DK T cells. At least 10,000 T cells were counted. (B) Coefficient of variation values for the DNA content within G1 peaks in cell-cycle profiles of Mps1DK/DK and Cre− T cells. Error bars show the SEM of more than five biological replicates for experimental animals or more than two replicates for control animals. (C) Kaplan–Meier curves showing overall survival of indicated genotypes. (D) Average mitotic index (% pH3) of thymocytes isolated from paclitaxel- or control-injected mice 4–6 h posttreatment. Error bars show SEM of more than five biological replicates in experimental animals or more than two replicates in control animals. (E) qPCRs showing complete loss of expression of p53 (p53 probes A and B) in lymphomas of indicated genotypes. Error bars show SEM for at least three tumors per genotype. (F) aCGH data showing the loss of the kinetochore-binding sequence in Mps1 in tumors with the indicated genotypes. Each rectangle represents a single aCGH probe value. Three probes values are shown: one probe recognizing the kinetochore-binding domain (Center) and two probes flanking the 5′ (Left) and 3′ (Right) sides of the deleted region. Log2 ratios less than −5 indicate complete loss of the indicated probe. Numbers on the left refer to tumor identification numbers (TID) (Dataset S1). (G) qPCRs showing full conversion of Mps1WT to Mps1DK in tumors with the indicated genotypes. Primer set A recognizes the sequence deleted in Mps1DK, and set B detects a fragment in the kinase domain. (H) Mps1 protein levels in p53f/f (TIDs 38 and 63 and full-length Mps1) and Mps1DK p53f/f tumors (TIDs 18 and 19) showing the conversion of full-length to truncated Mps1 in the latter genotype. The asterisk indicates a background band recognized in all lysates and runs just below Mps1DK that is detected only in TIDs 18 and 19.