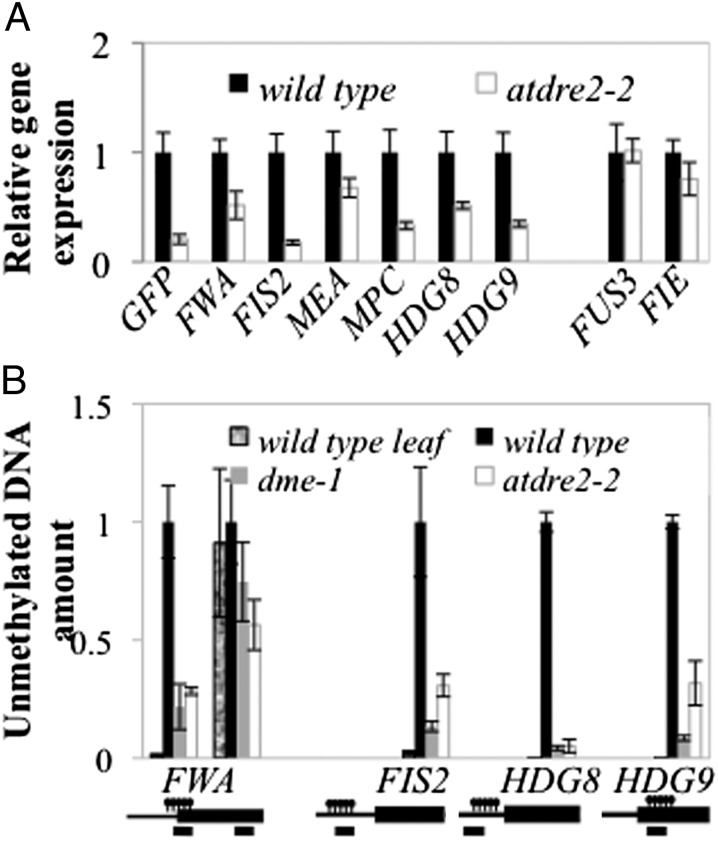
Fig. 4.
DNA methylation and gene expression defects in atdre2-2. (A) Gene expression of selected DME targets measured by RT-qPCR in 3 DAmP seeds dissected under a fluorescence microscope from pFWA-GFP and atdre2-2. (B) Estimation of endosperm DNA methylation levels from WT, dme-1, and atdre2-2 in 3 DAmP seeds. Equal amounts of seed DNA were digested with McrBC, and undigested genomic DNA was used as a standard for absolute quantification of specific regions. The leaf sample was used as a control for the digest. Promoters and genes are represented by black lines and boxes; the positions of methylated regions and PCR amplicons are indicated above and below, respectively. Values in A and B are mean ± SE from three or four biological replicates, normalized to UBIQUITIN10 (A) and an unmethylated region (B).