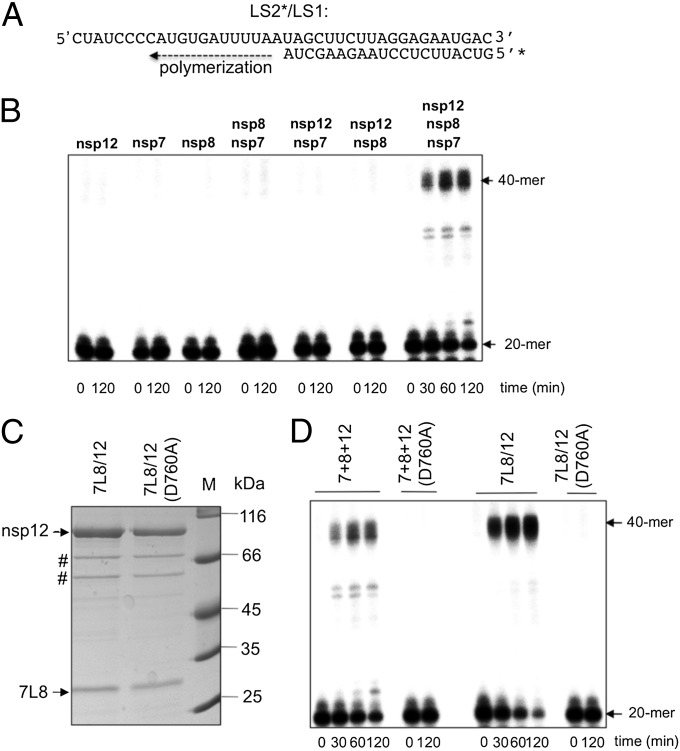
Fig. 1.
SARS-CoV nsp12 polymerase activity is activated by nsp7 and nsp8. (A) Sequence of the RNA primer/template used in this study; the 20-nt primer LS2 was 5′-radiolabeled (marked by *) and annealed to the 40-nt template LS1. (B) Primer extension polymerase assays were performed using LS2*/LS1 as substrate and different combinations of separately purified nsp12, nsp8, and nsp7. RNA products were separated by denaturing gel electrophoresis (20% polyacrylamide/7 M urea) and analyzed by autoradiography. The positions of the primer (20-mer) and the full-length extension product (40-mer) are indicated. (C) WT or mutant (D760A) nsp12 were coexpressed in E. coli with the nsp7-L-nsp8 fusion protein (7L8). After purification of the 7L8/nsp12 complex on a Strep-Tactin column, analysis by 12% SDS/PAGE and Coomassie blue staining of the proteins constituting the complex (nsp12 WT or D760A mutant) was done. # indicates the position of E. coli protein contaminants (defined by MALDI-TOF analysis). (D) Comparison of primer extension polymerase activities of WT and mutant (D760A) nsp12 in the presence of nsp7 and nsp8. Nsp7, nsp8, and nsp12 were either purified and added separately (lanes labeled 7+8+12) or copurified from E. coli as described above (lanes labeled 7L8/12). The reactions were performed on RNA template LS2*/LS1 (see B). Primer conversion rates (at 60 min): 40% for 7+8+12; 67% for 7L8/12; and 0% for 7+8+12(D760A) and 7L8/12(D760A).