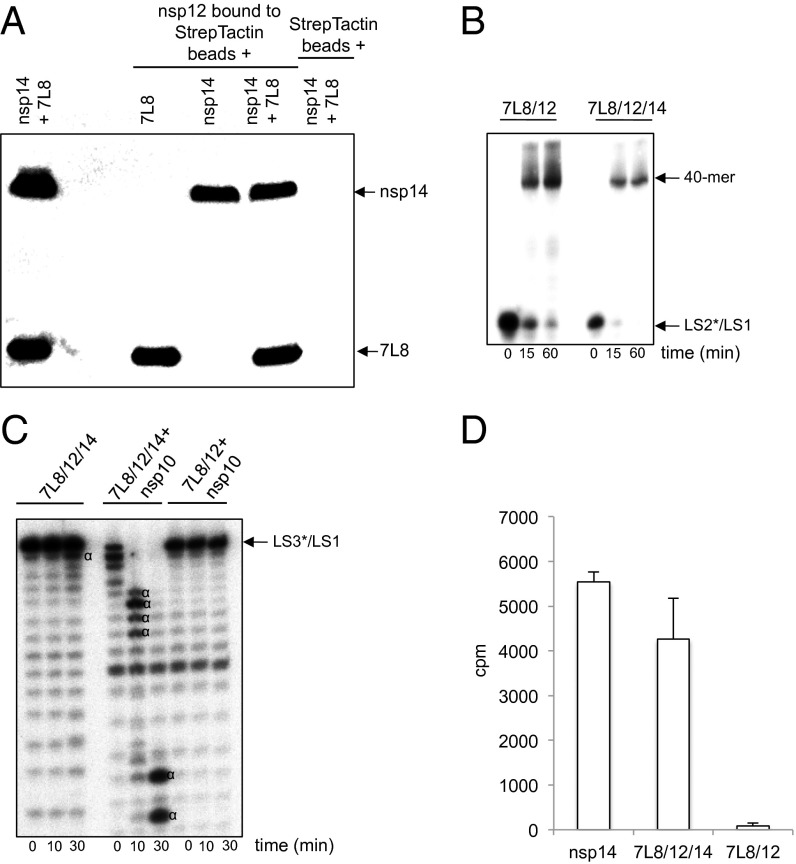
Fig. 6.
The SARS-CoV 7L8/12/14 complex possesses RdRp, ExoN, and N7-MTase activities. (A) Strep-tagged SARS-CoV nsp12 was bound to Strep-Tactin beads and incubated with 7L8, nsp14, or both simultaneously. After SDS/PAGE and Western blotting, his-tagged proteins (7L8 and nsp14) were revealed using an anti-His5-HRP antibody. (B) Time course primer extension polymerase assays were performed using either the 7L8/12 (500 nM) or the 7L8/12/14 (500 nM) complexes with LS2*/LS1 as primer*/template where LS2 was 5′-radiolabeled (marked by *). RNA products were separated in a denaturing polyacrylamide/urea gel and visualized by autoradiography. (C) Time course exoribonuclease assays were performed using the 7L8/12/14 (500 nM) complex in the absence or presence of 100 nM nsp10, and as control with 7L8/12 (500 nM) plus nsp10 (100 nM). The RNA substrate was a 40-nt RNA (LS1) annealed with 5′-radiolabeled LS3 primer carrying one noncomplementary base at its 3′ end (LS3*) and named LS3*/LS1. Digestion products were separated by denaturing polyacrylamide/urea gel electrophoresis and visualized by autoradiography (Fuji). The “α” symbol indicates RNA cleavage products. (D) AdoMet-dependent N7-MTase activity of the 7L8/12/14 complex. The different purified proteins or protein complexes (nsp14, 300 nM; 7L8/12/14, 300 nM; and 7L8/12, 300 nM) were incubated with substrate GpppAC4 RNA oligonucleotide in the presence of [3H]AdoMet. The methyl transfer to the capped RNA substrate was determined by using a filter-binding assay (as described in ref. 73). All experiments were done in triplicate (SDs are presented).