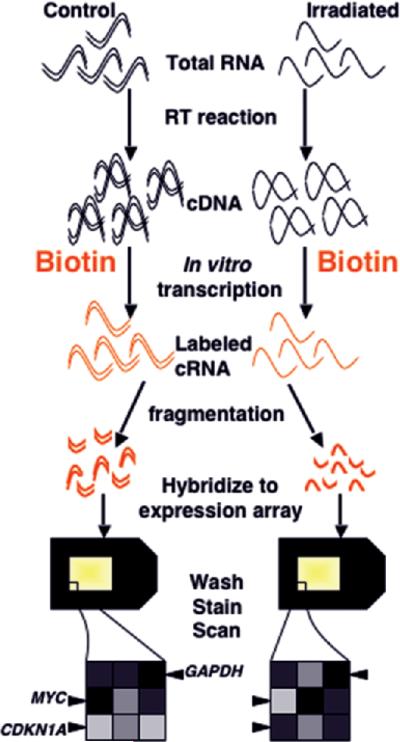
Figure 3.
Single-color microarray hybridization (photolithographic platform). Ribonucleic acid from the samples to be compared is reverse transcribed to complementary DNA (cDNA), then in vitro transcription is carried out in the presence of labeled (in this case bio-tinylated) nucleotides to produce labeled cRNA. The labeled complementary RNA (cRNA) is fragmented to facilitate sequence-specific hybridization, and each sample is hybridized to a separate array. After washing, staining, and scanning, the fluorescent intensity of each feature is compared between arrays, making normalization across experiments extremely important. In the illustrated experiment, features representing genes upregulated by radiation exposure, such as CDKN1A, will have a greater intensity on the array hybridized to the irradiated sample. Downregulated genes, such as MYC, will show a greater intensity in control samples, and unchanged genes, such as GAPDH, will have equal intensity on the two arrays.