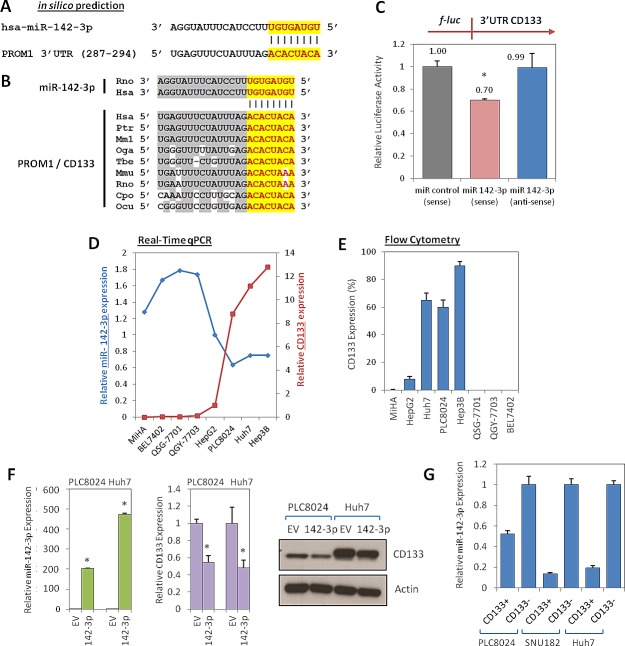
Figure 1. Regulation of CD133 by miR-142-3p.
(A) In silico prediction identified CD133 to be a target of miR-142-3p. (B) The chimpanzee (ptr: pan troglodytes), monkey (mml: macaca mulatta), bushbaby (oga; otolemur garnetti), treeshrew (tbe: tupaia belangeri), mouse (mmu: mus musculus), rat (rno: rattus norvegicus), guinea pig (cpo: cavia porcellus), rabbit (ocu: oryctolagus cuniculus) and human (hsa: homo sapiens) miR-142-3p and the predicted binding site in the 3'UTR of CD133 in different species were checked for alignment. The positions of the miRNA binding sites corresponded to the location of the GenBank sequence NM_006017. (C) Validation of miR-142-3p binding to the 3'UTR of CD133 by lucifearse reporter assay. Full length 3'UTR of CD133 (full-length) was PCR amplified and then cloned downstream of a firefly luciferase gene. Specificity of the inhibition was determined using anti-sense constructs (3' to 5' orientation). The pRL-TK Renilla luciferase plasmid was co-transfected as a normalization control for firefly luciferase activity. (D) The inverse correlations of CD133 (red line) and miR-142-3p (blue line) expression were determined across a panel of liver cell lines by qPCR. HepG2 was used as the calibrator. (E) Flow cytometry analysis of glycosylated protein CD133 expression in the same panel of liver cell lines. (F) Left panel: Stable overexpression of miR-142-3p in HCC cells PLC8024 and Huh7 by lentiviral transduction was confirmed by qPCR analysis. Middle and right panels: qPCR and Western blot analyses of CD133 expression following stable transduction of miR-142-3p or empty vector (EV) control in HCC cell lines PLC8024 and Huh7. (G) qPCR analysis for miR-142-3p expression in sorted CD133+ and CD133- subsets isolated from HCC cells PLC8024, SNU182 and Huh7.