SUMMARY
In organellogenesis of the chloroplast from endosymbiotic cyanobacterium, the establishment of protein targeting mechanisms to the chloroplast should have been pivotal. However, it is still mysterious how these mechanisms were established and how they work in plant cells. Here, we show that AKR2A, the cytosolic targeting factor for chloroplast outer membrane (COM) proteins, evolved from the ankyrin repeat domain (ARD) of the host cell by stepwise extensions of its N-terminal domain, and two lipids monogalactosyldiacylglycerol (MGDG) and phosphatidylglycerol (PG) of the endosymbiont were selected to function as the AKR2A receptor. Structural analysis, molecular modeling and mutational analysis of the ARD identified two adjacent sites for coincidental and synergistic binding of MGDG and PG. Based on these findings, we propose that the targeting mechanism of COM proteins was established using components from both the endosymbiont and host cell through a modification of the protein-protein interacting ARD into a lipid binding domain.
INTRODUCTION
Chloroplasts, an organelle responsible for photosynthesis in plants and algae (Dyall et al., 2004), evolved monophyletically from an ancient photosynthetic prokaryote cyanobacterium (Reyes-Prieto et al., 2007). The organellogenesis of the cyanobacterium into the chloroplast is thought to be accompanied by a massive transfer of genetic information from the endosymbiont to the host nucleus. Thus, although the chloroplast retains a functional genetic system of the endosymbiont, its genome was greatly reduced in size and now typically encodes only about 100 different proteins (Martin et al., 2002; Timmis et al., 2004). However, approximately 3,000 different proteins are required to build a fully functional chloroplast in plant cells. The majority of them are encoded by the nuclear genome and targeted from the cytosol to the chloroplasts after translation (Keegstra and Cline, 1999).
Chloroplast proteins can be largely grouped into two different categories; those imported into chloroplasts with the N-terminal transit peptide as a targeting signal and those targeted to chloroplast outer membrane (COM) without any cleavable signal sequence (Hofmann and Theg, 2005). Studies on the mechanisms of protein targeting to chloroplasts have mainly focused on how transit peptide-containing precursor proteins cross the chloroplast envelope membranes (Chen and Schnell, 1999). Many import factors and the mechanism of their actions in protein import into chloroplasts have been elucidated at the molecular and biochemical levels (Flores-Pérez and Jarvis, 2013; Kim and Hwang, 2013). However, the mechanisms by which proteins are targeted to and inserted in COM still remain poorly understood.
In the targeting of organellar proteins from the cytosol to their cognate organelles, the cytosolic targeting factor and its organelle-localized receptor constitute the key components of the targeting machinery as exemplified by the signal recognition particle (SRP) and its endoplasmic reticulum (ER)-localized SRP receptor (SR) for targeting of proteins to the ER (Keenan et al., 2001), and Pex5p and the peroxisomal docking complex (consisting of Pex14p, Pex13p and Pex17p) for targeting of PTS1-containing proteins to peroxisomes (Girzalsky et al., 2010). In Arabidopsis, ankyrin repeat protein 2A (AKR2A) and its close homolog AKR2B were identified as a cytosolic factor for targeting signal-anchored (SA) and tail-anchored (TA) proteins to the COM (Bae et al., 2008; Dhanoa et al., 2010). AKR2A specifically recognizes the targeting signals, the transmembrane domain (TMD) and the C-terminal positively charged flanking region (CPR), of client proteins through its N-terminal region and delivers them to the COM. Also, both AKR2A/B displays chaperone activity for COM-targeted SA and TA proteins and prevents non-specific aggregation of client proteins by binding to the hydrophobic TMD. In addition, AKR2A has the ability to bind to chloroplasts through its C-terminal ankyrin repeat domain (ARD). Another protein sHsp17.8, a member of the cytosolic class I small heat shock protein (sHsp) family, is also involved in the AKR2Amediated targeting of COM proteins. sHsp17.8 as a dimer binds to both AKR2A and chloroplasts. Through these interactions, sHsp17.8 facilitates the AKR2A-mediated targeting of SA proteins to chloroplasts (Kim et al., 2011). Despite this information on AKR2A and sHsp17.8, the exact mechanism by which AKR2A mediates protein targeting to chloroplasts remains largely unknown. In particular, it is not known how AKR2A recognizes chloroplasts as the target organelle and how the targeting mechanism evolved during organellogenesis of chloroplasts from the endosymbiotic cyanobacterium.
To address these fundamentally important questions, we investigated the identity of AKR2A receptor on the chloroplast surface and their mechanism of receptor recognition and the evolutionary origin of AKR2A. Our results show that AKR2A, which evolved from the ARD of the host cell, coincidentally and synergistically recognizes the two endosymbiont-derived lipids MGDG and PG as an AKR2A receptor for protein targeting to chloroplasts and, thereby, specifically delivers cargo proteins to COM.
RESULTS
AKR2A specifically binds to two chloroplast lipids, MGDG and PG
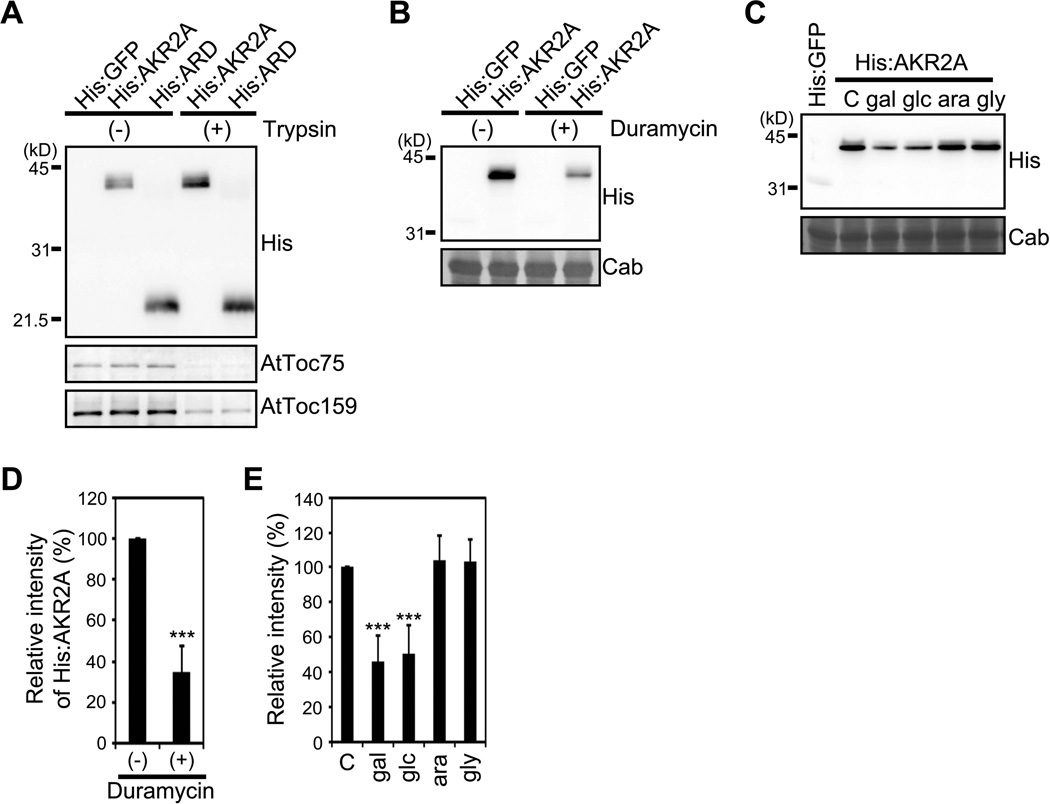
To elucidate the detailed mechanism of how AKR2A mediates specific protein targeting to chloroplasts, we identified a receptor of AKR2A. One possibility is that AKR2A interacts with a protein factor(s) on the surface of chloroplasts as the receptor for the targeting. Since a previous study showed that the ARD of AKR2A has binding ability to chloroplasts (Bae et al., 2008; Kim et al., 2011), we first defined the minimal chloroplast binding domain of AKR2A using an in vitro chloroplast binding assay using purified chloroplasts and various bacterially expressed AKR2A deletion mutants. The first three ankyrin repeats of AKR2A were sufficient for chloroplast binding (Figure S1A to S1C). To check the presence of a potential protein factor on the surface of chloroplasts for AKR2A binding, we examined whether the AKR2A binds to trypsin-treated chloroplasts. Trypsin can degrade proteins localized to the COM as well as to the intermembrane space between the outer and inner membranes (Jackson et al., 1998). Despite a significant reduction in the levels of two COM proteins AtToc75 and AtToc159 by the trypsin treatment (Figure 1A), binding of neither His:AKR2A nor His:ARD to chloroplasts was altered. We also examined if this interaction is mediated by small heat shock protein 17.8 (sHsp17.8), a cofactor of AKR2A (Kim et al., 2011). As reported previously (Kim et al., 2011), His:sHsp17.8 augmented AKR2A binding to chloroplasts (Figure S1D). Trypsin treatment of His:sHsp17.8-containing chloroplasts abrogated this augmentation but did not affect the intrinsic chloroplast binding activity of AKR2A, showing that sHsp17.8 is not essential for chloroplast binding of AKR2A. Collectively, these results suggest that AKR2A recognizes a non-protein factor for initial chloroplast binding.
Figure 1. AKR2A recognizes the lipid components of chloroplasts for its binding.
(A) The effect of trypsin treatment on AKR2A binding to chloroplasts. His:AKR2A was incubated with chloroplasts treated with (+) or without (−) trypsin and the amount of His:AKR2A copurified with chloroplasts was determined by Western blot analysis using anti-His, anti-AtToc75 and anti-Toc159 antibodies. The amount of AtToc75 and AtToc159 was analyzed as internal control for trypsin treatment. (B–E) Effect of duramycin and sugars on AKR2A-chloroplast binding.
(B and C) AKR2A binding to chloroplasts treated with (+) or without (−) duramycin (B) or in the presence of the indicated sugars (C). (B and C) Western blots analysis of AKR2A binding to chloroplasts using anti-His antibody.
(D and E) quantification of AKR2A binding to chloroplasts. Mean ± standard deviation (SD) are shown (n = 3). C, control (no sugar); glc, glucose; gal, galactose; gly, glyceraldehyde; ara, arabinose. The asterisks indicate a significant difference from the corresponding control experiment by Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001). See also Figure S1.
Many proteins are targeted to subcellular membranes by binding to particular lipids enriched in the membranes, such as phosphatidylinositol-4,5-bisphosphate in the plasma membrane (McLaughlin et al., 2002; Yoon et al., 2011). Chloroplasts contain unique lipids, such as MGDG and digalactosyldiacylglycerol (DGDG) (Block et al., 1983). Moreover, these chloroplast-specific lipids are thought to play a role in protein targeting (Schleiff et al., 2001). To test whether lipids are involved in the binding of AKR2A to chloroplasts, we examined the effect of treating chloroplasts with duramycin, which causes aggregation of phosphatidylethanolamine (PE) and MGDG in membranes (Navarro et al., 1985), on His:AKR2A binding to chloroplasts. The duramycin treatment significantly reduced the chloroplast binding of His:AKR2A (Figure 1B and 1D). Because COMs contain no significant amount of PE (Block et al., 1983), this result suggests that MGDG is important for the AKR2A binding to chloroplasts. To corroborate this idea, we examined whether galactose, the sugar moiety of MGDG, and other related compounds inhibit the His:AKR2A binding to chloroplasts. Among the compounds we tested, two hexoses, galactose and glucose, reduced the His:AKR2A binding to chloroplasts (Figure 1C and 1E). However, glyceraldehyde and arabinose did not affect the binding, indicating that AKR2A recognizes hexoses.
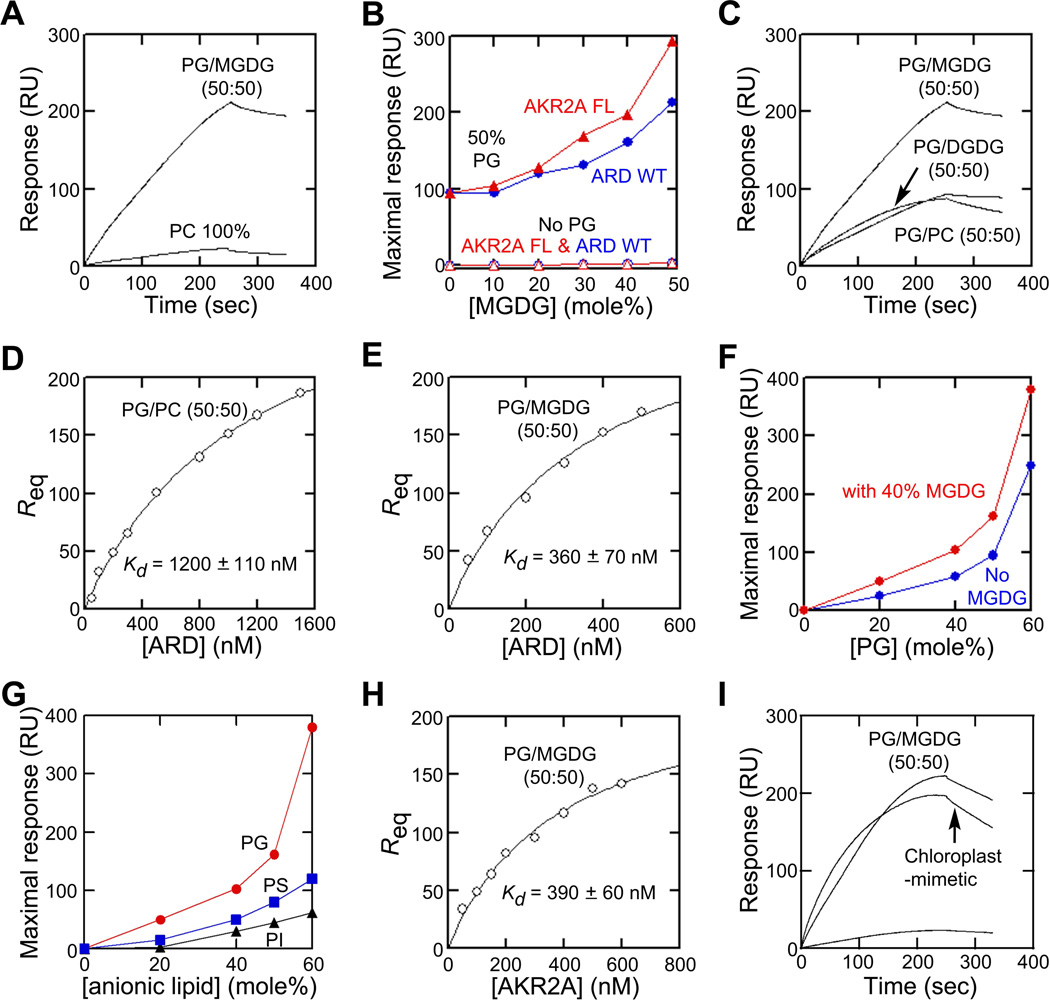
To confirm that AKR2A specifically binds MGDG, we measured the binding of His:AKR2A and His:ARD to the vesicles of various lipid compositions by surface plasmon resonance (SPR) analysis. His:ARD exhibited negligible binding to the zwitterionic phospholipid phosphatidylcholine (PC) (Figure 2A); thus, we used PC as an inert bulk lipid in the vesicles. We found that His:ARD displayed little binding to the vesicles containing PC and MGDG (Figure 2B), showing that the ARD cannot effectively bind MGDG alone. However, the inclusion of a chloroplast-abundant lipid, PG, allowed His:ARD to bind vesicles in an MGDG-dependent manner; when His:ARD was added to vesicles with a fixed PG concentration and varying MGDG concentrations (i.e., POPG/POPC/MGDG [50:(50−x):x in mole%]), binding increased monotonously with increasing MGDG concentrations (Figure 2B). Under the same conditions, His:ARD showed much lower binding to another chloroplast-specific lipid, DGDG, demonstrating that the MGDG binding is specific (Figure 2C).
Figure 2. Membrane binding properties of AKR2A and ARD.
(A) ARD shows negligible binding to PC vesicles when compared with PG/MGDG (50:50) vesicles.
(B) MGDG-dependent vesicle binding of full-length (FL) AKR2A (red) and ARD (blue) with POPC/MGDG [(100−x):x; x, mole% of MGDG] (open symbols) or POPG/POPC/MGDG [50:(50−x):x] (closed symbols) vesicles and maximal binding response values plotted against MGDG concentration.
(C) Specificity of ARD for MGDG over DGDG. Notice that binding to PG/DGDG (50:50) vesicles is comparable to that of PG/PC (50:50) vesicles, showing that the affinity for DGDG alone is as low as that for PC.
(D) Determination of Kd for ARD binding to PG/PC (50:50) vesicles by equilibrium SPR analysis. The binding isotherm was generated from the response at equilibrium (Req)(average of triplicate measurements) versus the ARD concentration (P0) plot. A solid line represents a theoretical curve constructed from the Rmax (330 ± 20) and Kd (1200 ± 110 nM) values determined by a nonlinear least squares analysis of the isotherm using the following equation: Req = Rmax/(1 + Kd/P0).
(E) Determination of Kd for ARD binding to PG/MGDG (50:50) vesicles by equilibrium SPR analysis as described for Figure 3D. Rmax = 290 ± 30 and Kd = 360 ± 70 nM.
(F) PG-dependent vesicle binding of the ARD with POPC/POPG [(100−x):x] (blue) or POPC/POPG/MGDG [(60−x):x:40] (red) vesicles and maximal binding response values plotted against PG concentration.
(G) PG specificity of ARD. Binding of the ARD (0.5 µM) to PC/PG [(100−x):x] (red), PC/PS [(100−x):x] (blue) or PC/PI [(100−x):x] (black) vesicles was measured by kinetic SPR analysis, and maximal binding response values were plotted against anionic lipid concentrations.
(H) Determination of Kd for full-length AKR2A binding to PG/MGDG (50:50) vesicles by equilibrium SPR analysis as described for Figure 2D. Rmax = 230 ± 20 and Kd = 390 ± 60 nM.
(I) AKR2A showed significantly higher binding to chloroplast-mimicking vesicles (MGDG/DGDG/PC/PG/PI/sulfoquinovosyldiacylglycerol = 17:29:32:10:6:6) than to 100% PC vesicles and its binding to chloroplast-mimicking vesicles was comparable to that to PG/MGDG vesicles (50:50). For SPR data, each point represents the average of triplicate measurements (n = 3). RU, resonance unit.
We determined the Kd values of His:ARD for vesicles composed of POPG/POPC (50:50) and POPG/MGDG (50:50) (Figure 2D and 2E). This indicated that 50 mole% MGDG causes an approximately 3-fold increase in membrane affinity over PG alone. His:ARD also binds vesicles in a PG-dependent manner with or without MGDG; however, in the presence of 40% MGDG, His:ARD shows a sharper PG dependence (Figure 2F), suggesting that ARD binds PG and MGDG simultaneously and synergistically. Under the same conditions, other anionic lipids, phosphatidylserine (PS) and phosphatidylinositol (PI), were much less effective than PG in inducing membrane binding of His:ARD with or without MGDG (Figure 2G), suggesting a specific effect of PG on the binding. The full-length His:AKR2A showed similar membrane binding properties to His:ARD; it had essentially the same affinity (Kd = 390 ± 60 nM) for POPG/MGDG (50:50) vesicles (Figure 2H) as His:ARD and showed the same ARD-like linear dependency on MGDG in vesicle binding (Figure 2B). AKR2A also showed significantly higher binding to vesicles whose lipid composition mimics that of COM (MGDG/DGDG/PC/PG/PI/sulfoquinovosyldiacylgl ycerol = 17:29:32:10:6:6) (Block et al., 1983) than to 100% PC vesicles and its binding to chloroplastmimicking vesicles was comparable to that to PG/MGDG vesicles (50:50) (Figure 2I).
MGDG and PG binding of AKR2A is physiologically important
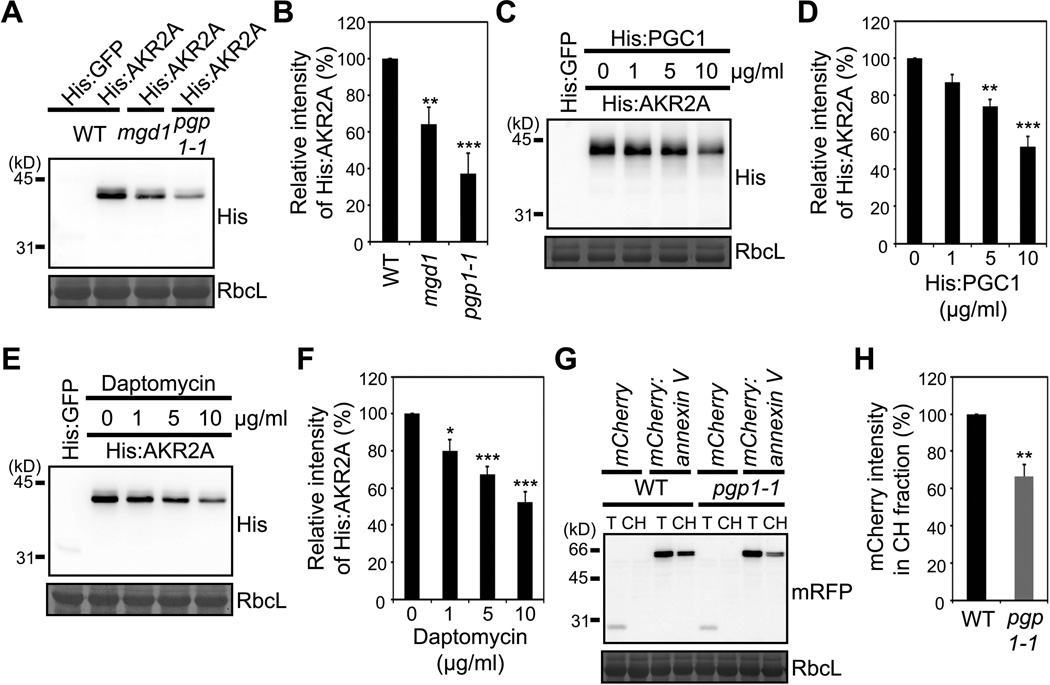
To examine the physiological significance of AKR2A binding to MGDG and PG, we measured the AKR2A binding to chloroplasts of monogalactosyldiacylglycerol synthase 1 (MGD1) and phosphatidylglycerophosphate synthase 1 (PGP1) mutants. MGD1 and PGP1 are involved in the production of MGDG and PG in the chloroplasts, respectively (Babiychuk et al., 2003; Jarvis et al., 2000; Kobayashi et al., 2007; Shimojima et al., 1997; Xu et al., 2002). Although the null mutants mgd1-2 and pgp1 are lethal (Babiychuk et al., 2003; Kobayashi et al., 2007), leaky mutants, mgd1 and pgp1-1, exhibit a yellow leaf phenotype due to lower MGDG and PG levels in the leaves (Jarvis et al., 2000; Xu et al., 2002). In the mgd1 mutant plants, the enzymatic activity of MGD1 was reduced to 75% of wild-type (WT) plants, which caused a 42% reduction in the amount of MGDG (Jarvis et al., 2000). In the case of pgp1-1 mutant plants, the PGP1 activity was decreased by 80%, which resulted in a 30% reduction in the overall PG content (Xu et al., 2002). Thus, we used chloroplasts from mgd1 and pgp1-1 for AKR2A binding. The amount of His:AKR2A bound to mgd1 or pgp1-1 chloroplasts was significantly lower than that bound to WT chloroplasts (Figure 3A and 3B). These results are consistent with the notion that MGDG and PG in COM are responsible for the COM binding of AKR2A. In case of PG, however, our data may be in conflict with the previous report by Dorne et al. (1985), suggesting that PG is mainly localized in the inner leaflet of the COM. We therefore performed further experiments to prove that PG in COM is available to the cytosolic AKR2A. First, we treated purified chloroplasts with recombinant His-tagged yeast PG-specific phospholipase C (His:yPGC1) (Simocková et al., 2008) and used them for AKR2A binding. The His:yPGC1 treatment reduced the chloroplast binding of His:AKR2A in a dose-dependent manner (Figure 3C, 3D, Figure S2A and S2B). We also examined the effect of daptomycin on the chloroplast binding of AKR2A. Daptomycin is a lipopeptide antibiotic that specifically binds to PG so as to form oligomers, and thereby can depolarize the cell membrane potential (Hachmann et al., 2009 and 2011; Muraih et al., 2011 and 2012). The daptomycin treatment reduced the chloroplast binding of His:AKR2A (Figure 3E and 3F), supporting the notion that PG is present in the outer leaflet of the COM. We then examined binding of annexin V to WT and pgp1-1 chloroplasts. Annexin V non-specifically binds to negatively charged phospholipids, including PS and PG, in a calcium-dependent manner (Jeppesen et al., 2008) and it should show different affinity for chloroplasts from WT and pgp1-1 plants if PG is exposed to the outer layer of COM. The COM is known to contain a negligible amount of PS (Block et al., 1983). Annexin V tagged with mCherry at the N-terminus was transiently expressed in pgp1-1 or WT protoplasts, and its binding to chloroplasts was determined by Western blot analysis using the purified chloroplast fraction. As shown in Figure 3G and 3H, the amount of mCherry:annexin V copurified with pgp1-1 chloroplasts was significantly lower than that of WT chloroplasts, supporting the notion that PG is present in the outer leaflet of the COM. Furthermore, we tested if the reduced AKR2A binding to mgd1 and pgp1-1 chloroplasts could be restored by exogenously added MGDG and PG, respectively. Specifically, liposomes were prepared from the envelope membranes of mgd1 and pgp1-1 chloroplasts that had been supplemented with MGDG and PG, respectively, and used for AKR2A binding in vitro. As shown in Figure S2C and S2D, supplementation of MGDG and PG, respectively, to the envelope membranes of mgd1 and pgp1-1 chloroplasts fully restored vesicle binding of His:AKR2A in a dose-dependent manner. Moreover, this restoration was abolished by the treatment of duramycin and daptomycin (Figure S2C and S2D). However, supplementation of PG to envelope membranes of mgd1 chloroplasts, and MGDG to envelope membranes of pgp1-1 chloroplasts did not restore or only marginally restored His:AKR2A binding to liposomes (Figure S2E). Also, binding of His:AKR2A to pgp1-1 chloroplasts could not be effectively rescued by PS and PI, further underscoring the specific nature of PG binding (Figure S2F and S2G). PS was slightly more effective than PI, which reflects modestly higher affinity of AKR2A for PS than for PI (see Figure 2G). Collectively, these results show that the reduced binding of AKR2A to mgd1 and pgp1-1 chloroplasts is the direct effect of the reduced MGDG and PG levels on the COM, respectively.
Figure 3. Chloroplast binding of AKR2A is impaired in mgd1 and pgp1-1 mutants.
(A and B) His:AKR2A binding to chloroplasts from WT, mgd1 or pgp1-1 plants. (A) Western blot analysis of His:AKR2A bound to chloroplasts using anti-His antibody. RbcL, loading control stained with Coomassie blue. (B) Quantification of His:AKR2A binding to mgd1, pgp1-1 or WT chloroplasts. Mean ± SD are shown (n = 3).
(C and D) The effect of the yPGC1 treatment to chloroplasts on AKR2A binding. (C) His:AKR2A binding to chloroplasts was examined as in (A) except that chloroplasts had been treated with His:yPGC1 before its use in binding experiments. (D) Quantification of His:AKR2A binding to His:yPGC1-treated chloroplasts. Mean ± SD are shown (n = 3).
(E and F) Effect of daptomycin on AKR2A binding to chloroplasts. (E) His:AKR2A binding to chloroplasts was examined as in (A) except that chloroplasts had been treated with daptomycin before its use in binding assay. (F) Quantification of His:AKR2A binding to daptomycin-treated chloroplasts. Mean ± SD are shown (n = 3).
(G and H) The binding of annexin V to chloroplasts. (G) mCherry:annexin V or mCherry alone was introduced into protoplasts and chloroplast fractions from the transformed protoplasts were analyzed by Western blotting using anti-RFP antibody. mCherry alone was used as a control for fractionation. T, total protoplast extracts; CH, chloroplast fractions. (H) Quantification of the chloroplast-bound mCherry:annexin V. Mean ± SD are shown (n = 3). The asterisks indicate a significant difference from the corresponding control experiment by Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001). See also Figure S2.
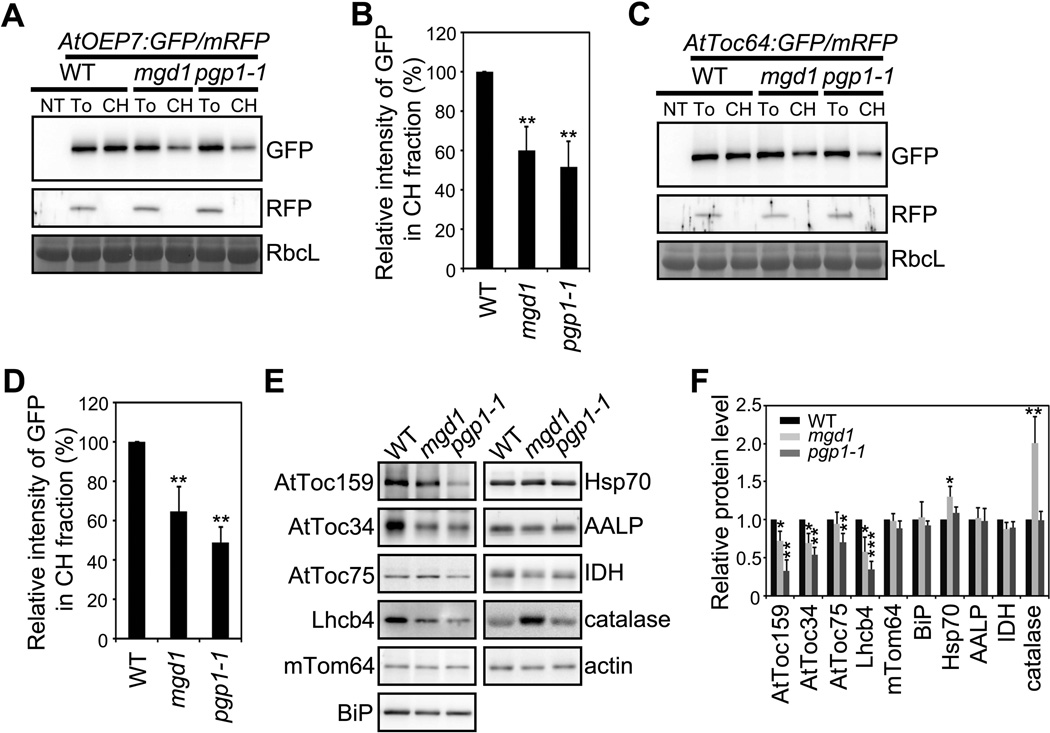
We examined whether the lower MGDG and PG levels in chloroplasts affect the protein targeting to chloroplasts. GFP-tagged COM protein constructs AtOEP7:GFP, AtToc64:GFP or GFP:AtToc34 (Dhanoa et al., 2010; Lee et al., 2001; Lee et al., 2004) were cotransformed into mgd1, pgp1-1 or WT protoplasts, and their chloroplast targeting efficiency was determined (Figure 4A, 4C and Figure S3A). The amount of these proteins copurified with mgd1 or pgp1-1 chloroplasts was lower than that of WT plants (Figure 4B, 4D and Figure S3B). To further assess the effect of these mutations on protein targeting to chloroplasts, we measured the levels of various organellar proteins in mgd1, pgp1-1 and WT plants using specific antibodies (Figure 4E and 4F). Compared with WT plants, mgd1 and pgp1-1 plants had significantly reduced levels of chloroplast proteins, with the exception of AtToc75 in mdg1 plants (Figure 4E and 4F). The lack of effect on the level of AtToc75 in mdg1 chloroplasts might be due to the fact that the transcript level of AtToc75 was increased by 40% in mgd1 plants (Figure S3C). In contrast, the levels of all other proteins, including cytosolic actin and Hsp70 (in pgp1-1), ER BiP, vacuolar AALP, and mitochondrial mTom64 and IDH proteins, were not affected (or higher for peroxisomal catalase) in these mutants. These results indicate that MGDG and PG play a specific and pivotal role in the targeting of chloroplast proteins.
Figure 4. Targeting of proteins to the COM is impaired in mgd1 and pgp1-1 mutants.
(A–D) The targeting efficiency of COM proteins in mgd1 and pgp1-1 mutants. (A and C) Targeting of AtOEP7:GFP (A) or AtToc64:GFP (C) to chloroplasts in WT, mgd1 or pgp1-1 protoplasts. mRFP, a control for the transformation efficiency and chloroplast fractionation. RbcL, loading control. NT, non-transformed; To, total protoplast extracts; CH, chloroplast fractions. (B and D) Quantification of the chloroplast targeting of AtOEP7:GFP (B) or AtToc64:GFP (D). Mean ± SD are shown (n = 3).
(E and F) Chloroplast protein levels in mgd1 and pgp1-1 mutants. (E) Western blot analysis of various endogenous protein levels. Actin, loading control. (F) Quantification of the protein levels in mgd1 and pgp1-1 plants. The protein levels were normalized using Actin. The expression level in WT plants was set to 1. Mean ± SD are shown (n = 3). The asterisks indicate a significant difference from the corresponding control experiment by Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001). See also Figure S3.
Structural basis for the AKR2A binding to MGDG and PG
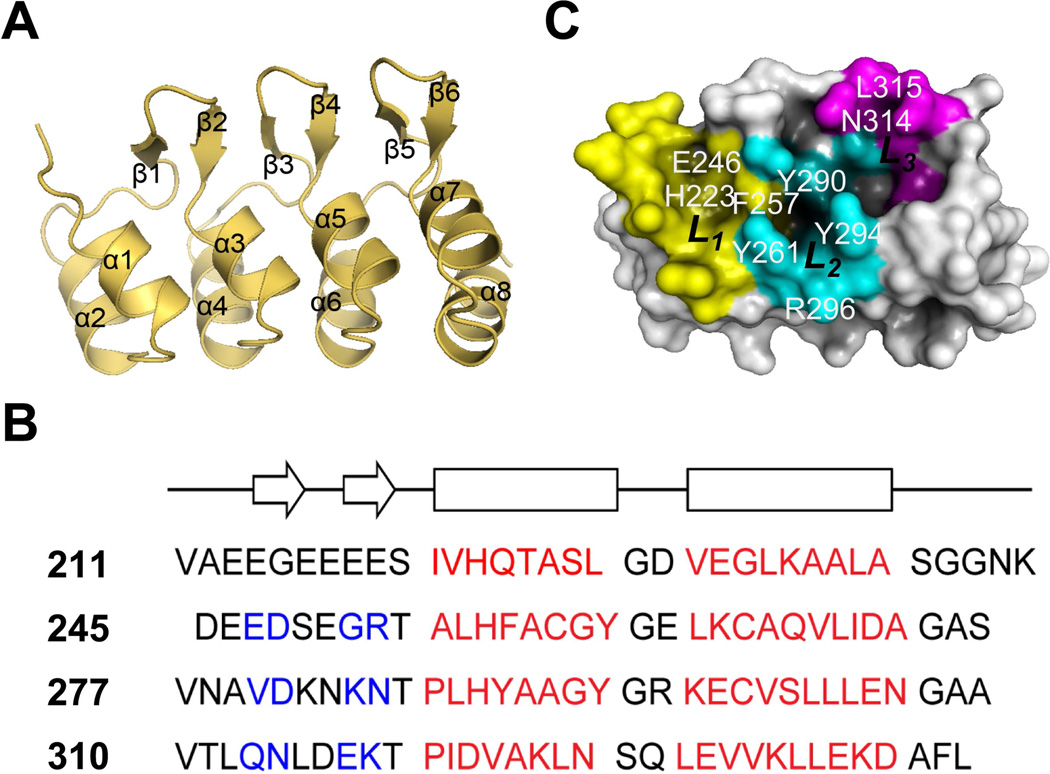
To understand the structural basis for the AKR2A binding to MGDG and PG, we determined the crystal structure of the ARD of AKR2A at 2.3 Å resolution (Figure 5A and Table S1). Each of four ankyrin repeats has an anti-parallel helix-turn-helix, a β-hairpin (Figure 5B) and the four repeats stacked together to form an L-shaped domain. The structural determination of the MGDG and PG binding sites by the co-crystallization of ARD with galactose and glycerol, respectively, was unsuccessful because of the low binding affinity of ARD for these lipid headgroups. This low affinity is not surprising, because optimal lipid binding normally requires lipids in the membrane environment (Cho and Stahelin, 2005).
Figure 5. The overall structure of ARD.
(A) The crystal structure ARD of AKR2A at 2.3 Å resolution.
(B) Four ankyrin repeats of AKR2A are aligned according to their structure. Arrows and rectangles indicate the approximate locations of the β-strands (blue) and α-helices (red), respectively.
(C) The putative membrane binding surface of the ARD. There are three grooves (L1, L2, and L3 in yellow, cyan, and magenta, respectively, with key residues constituting each site indicated) that can accommodate lipid headgroups. See also Table S1.
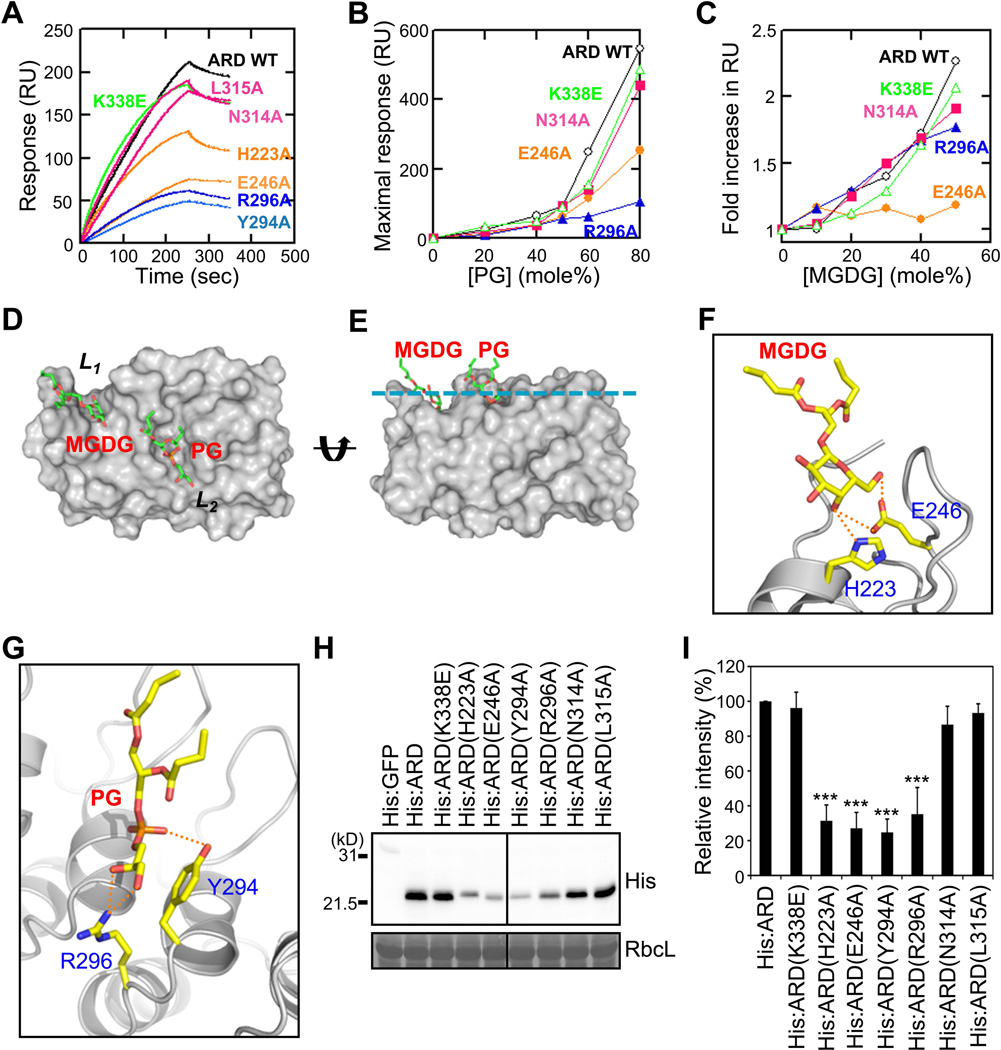
In most lipid binding proteins, their lipid binding surfaces contain a cluster of aromatic and basic (or other polar) residues that can interact either non-specifically with the membrane surface or specifically with the lipid headgroup (Chen et al., 2012; Sumandea et al., 1999). Consistent with the lipid binding activity of AKR2A, surface analysis of its ARD revealed that one face of this domain has many exposed aromatic residues and three grooves that can accommodate lipid headgroups. One of these grooves (L1) is formed with the residues H223, Q224, S227, E246, D248 and F257. The second pocket (L2) is composed of F257, Y261, Y290, Y294 and R296, and the third one (L3) contains N314, L315, L326 and N327 (Figure 5C). To understand the lipid binding mode of ARD, we modeled the lipid binding of MGDG and PG to ARD. This analysis suggested that MGDG binds to L1, whereas PG binds to L2 or L3. We examined the lipid binding of ARD to these grooves by mutating some of the residues that were predicted to interact with the lipid headgroups in the model, particularly those conserved in plant AKR2A homologs (Figure S4), and then measuring the effects of these mutations on ARD binding to vesicles. Toward POPG/POPC (50:50) vesicles, the L2 site mutants (Y294A and R296A) showed significantly lower binding than WT, whereas the L3 site mutants (N314A and L315A) showed only modestly reduced binding (Figure 6A). Additionally, a representative L2 site mutant (R296A) showed a lower PG dependency than WT in vesicle binding, while a L3 site mutant (N314A) exhibited a WT-like PG dependency (Figure 6B). Furthermore, the R296A mutant had essentially the same MGDG concentration dependency as WT (Figure 6C). These data suggest that the L2 site residues are involved in PG binding, but not in MGDG binding, whereas the L3 site residues are not involved in lipid binding. The L1 site mutants H223A and E246A also had a significantly lower binding affinity for POPG/POPC (50:50) vesicles than WT (Figure 6A). Unlike the L2 site mutants, however, a representative L1 mutant, E246A, showed some PG dependence (Figure 6B) and very low MGDG dependence in vesicle binding (Figure 6C). These results suggest that the L1 site residues are directly involved in the MGDG binding and that PG can also occupy the L1 site in the absence of MGDG. The K338E mutant, a negative control, had WT-like vesicle binding (Figure 6A, 6B and 6C).
Figure 6. Identification of the MGDG- and PG-binding sites of the AKR2A ARD.
(A) SPR sensorgrams of ARD mutants interacting with POPC/POPG (50:50) vesicles.
(B) Measurement of the PG-dependent vesicle binding of the ARD mutants with POPC/POPG [(100−x):x] vesicles and maximal binding response values plotted against PG concentration.
(C) The MGDG-dependent vesicle binding of the ARD mutants. Maximal response values for ARD mutants interacting with POPG/POPC/MGDG [50:(50−x):x] vesicles measured at each MGDG concentration. Mutants in each group (L1, L2 and L3) show similar properties; thus, data for a representative mutant for each group (E246A for L1, R296A for L2, and N314A for L3) are shown in B and C for clarity. L1, L2, L3 site mutants are represented by orange, blue, and magenta, respectively. For SPR data, each point represents the average of triplicate measurements. RU, resonance unit. n = 3.
(D and E) A modeled structure of the ARD-PG-MGDG complex in two different orientations. (D) The ARD is shown in the same molecular orientation as in Figure 6C. (E) The structure is horizontally rotated 90° to show its membrane binding orientation. The cyan line indicates the putative membrane surface.
(F) Predicted hydrogen bonds between the MGDG headgroup and two key residues, H223 and E246, in the L1 pocket are shown as green dotted lines.
(G) Potential hydrogen bonds between the PG headgroup and two key residues, Y294 and R296, in the L2 pocket are shown as green dotted lines. The ternary complex model is identical to that shown in Figure 6D and 6E. The molecular orientations are arbitrarily selected for the best illustration of potential interactions. Although two key residues are shown for each site, many other protein residues can also participate in short-range interactions with lipid headgroups and acyl chains. Lipids are in stick representation and proteins in ribbon representation.
(H) Chloroplast binding of ARD and L1, L3, and L3 site mutants (see Figure 5C). RbcL, loading control.
(I) Chloroplast binding of L1, L3, and L3 site mutants was presented as relative values to that of His:ARD. Mean ± SD are shown (n = 3). The asterisks indicate a significant difference from the corresponding control experiment by Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001). See also Figure S4.
On the basis of these mutational data, we propose a molecular model of the ARD-PG-MGDG complex (Figure 6D to 6G). Optimal binding is achieved when the L1 and L2 sites are occupied by MGDG and PG, respectively. Our results also suggest that the simultaneous binding of PG and MGDG to their respective sites is necessary for tighter and more specific binding. We measured the chloroplast binding of the ARD and its mutants to test if those mutants with compromised lipid binding show reduced chloroplast binding. Figure 6H and 6I shows that there is an excellent correlation between lipid binding and chloroplast binding of the ARD. Collectively, these results show that two adjacent grooves are involved in binding to PG and MGDG and that this lipid binding is a driving force for the chloroplast binding of AKR2A.
AKR2A evolved by a stepwise addition of the N-terminal domains to the ARD
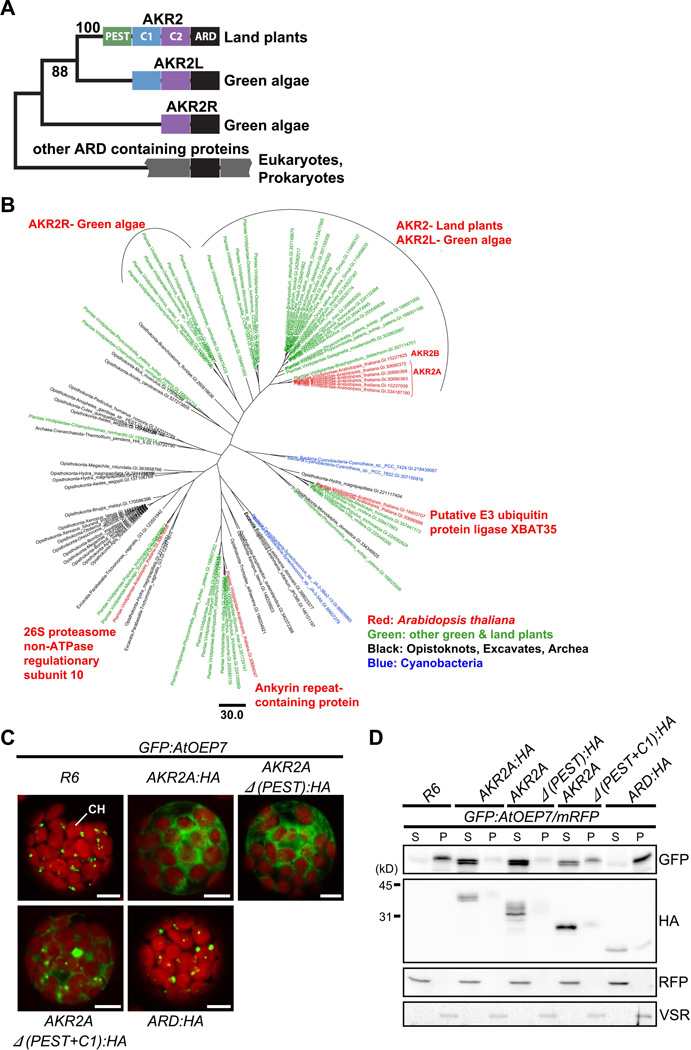
To gain insight into how the chloroplast protein targeting mechanisms were established during organellogenesis of the chloroplast, we examined the evolutionary origin of the cytosolic targeting factor, AKR2A, using a phylogenetic approach. AKR2A contains four conserved regions, the PEST sequence (Rechsteiner and Rogers, 1996), two central domains (C1 and C2) and C-terminal ARD (Figure 7A). Among these four regions, the ARD was highly conserved in land plants and certain green algae but displayed a clear phylogenetic separation from that of cyanobacteria, the progenitor of chloroplasts (Figure 7B and Figure S5), raising the possibility that AKR2A evolved from the ARD of the host cell. The large number of AKR2-type ARD-containing proteins in green algae and land plants were classified into three groups based on the N-terminal domain composition. Of these groups, AKR2-related (AKR2R) proteins contain only the C2 as the N-terminal domain and exist in some green algae (Volvox, Chlamydomonas, Micromonas, and Ostreococcus) (Figure 7A and Figure S5) whereas AKR2-like (AKR2L) proteins have both C1 and C2 domains and also present in certain green algae, raising the possibility that the C1 domain was added to the N-terminus of AKR2L after gene duplication but prior to divergence of land plants (Finet et al., 2010). The third group, AKR2, contains all three N-terminal parts and exists only in land plants (mosses, ferns, and angiosperms), suggesting that the PEST sequence was added to AKR2L after the evolution of land plants from green algae (Figure 7A). These raised the possibility that AKR2L and AKR2R are the evolutionary intermediates of AKR2 in plants.
Figure 7. The Phylogenetic tree of ARDs and the evolution of AKR2A.
(A) The domain structure of ARD-containing proteins. The PEST (green), C1 (blue), C2 (purple) and ARD domains are highlighted in different colors. In cyanobacterial ARD-containing proteins, only the ARD domain (black) is shown.
(B) Maximum likelihood phylogenetic tree of ARDs. The tree is built on an alignment of 115 amino acid residues of the ARDs of 93 sequences.
(C and D) The effect of C1 and C2 domains on AKR2A binding to GFP:AtOEP7. (C) GFP:AtOEP7 was introduced into protoplasts together with HA-tagged full-length or various deletion mutants of AKR2A or empty vector R6 and the localization pattern of GFP:AtOEP7 was examined. CH, chloroplasts. Bar, 10 µm.
(D) Fractionation of GFP:AtOEP7. Protoplasts were transformed with GFP:AtOEP7 and AKR2A as in (C). mRFP was included in all transformation as a control for transformation efficiency and fractionation. Protoplast lysates were separated into soluble and pellet fractions and analyzed by Western blotting using anti-GFP, anti-HA, anti-RFP and anti-VSR antibodies. VSR was used as a control for membrane proteins. S, soluble fraction; P, pellet fraction. See also Figure S5.
The N-terminal region of AKR2A is involved in cargo binding (Bae et al., 2008). Thus, the stepwise extension of each part of the N-terminal region may have a certain relationship with the cargo binding ability. To test this idea, we generated three AKR2A mutants that had a deletion of the PEST sequence alone ((AKR2AΔ(PEST)), the PEST sequence plus the C1 domain ((AKR2AΔ(PEST+C1)), or all three N-terminal parts ((AKR2AΔ(PEST+C1+C2)), to mimic AKR2L, AKR2R and ARD, respectively, in the domain structure, and examined their cargo binding ability. The binding of AKR2A to its cargo protein, AtOEP7, can be visualized using GFP:AtOEP7 (Bae et al., 2008). The fusion of GFP to the N-terminus of AtOEP7 inhibits its targeting to chloroplasts and results in its aggregation in the cytosol. However, coexpression of AKR2A solubilizes GFP:AtOEP7 in the cytosol because AKR2A binds to the hydrophobic TMD of GFP:AtOEP7. Similar to AKR2A:HA, AKR2AΔ(PEST):HA solubilized GFP:AtOEP7 (Figure 7C and 7D). In contrast, AKR2AΔ(PEST+C1):HA and AKR2AΔ(PEST+C1+C2):HA solubilized GFP:AtOEP7 only partially or not at all, respectively, suggesting that AKR2Ls and AKR2Rs have partial and no cargo binding activity, respectively. In addition, AKR2AΔ(PEST):HA was prone to degradation. These results raised the possibility that the cargo binding ability of AKR2A evolved by a stepwise addition of C1 and C2 to the ARD to give AKR2R and AKR2L, and that the PEST sequence was added to regulate the stability of AKR2s after plant evolution.
DISCUSSION
This study focuses on three aspects of protein targeting to the COM; how AKR2A, the cytosolic targeting factor of COM proteins, developed during evolution, what is the chloroplast-localized receptor for AKR2A, and finally what is the mechanism of the interaction between AKR2A and its receptor. Our results provide important clues to all these fundamentally important questions and provide new insight into how the targeting mechanism was established during evolution.
The mechanism of the AKR2A-mediated protein targeting to chloroplasts
The 33-amino acid ankyrin repeat is one of the most common protein-protein interaction motifs that is found in nearly 6% of all eukaryotic protein sequences. This small module is known to be involved in diverse cellular functions, including cytoskeletal organization, cell signaling, transcriptional regulation, inflammatory responses, cell cycle regulation, cell development, and cell differentiation, by mediating specific protein-protein interactions (Barrick et al., 2008; Mosavi et al., 2004). However, the lipid binding activity of the ankyrin repeat has not been reported. In this study, we provide compelling new evidence that the ankyrin repeat can also serve as a lipid binding module, joining the list of expanding families of lipid binding protein-interaction domains (Chen et al., 2012; Sheng et al., 2012). The number of ankyrin repeats found in a single protein varies widely depending on proteins, but on average two to four repeats form an ARD. The four ankyrin repeats in AKR2A exhibit the typical helix-turn-helix conformation. Although the overall ARD structure of AKR2A is similar to a canonical ARD structure (Li et al., 2006; Mosavi et al., 2004), it has many exposed aromatic residues and well-formed grooves on the same surface that have not been described in other ARDs, both of which are features important for effective membrane-protein interactions (Cho & Stahelin 2005).
The ARD specifically binds to PG and MGDG, two abundant lipids in chloroplasts, for chloroplast binding in vitro. In addition, other factors may play a role in AKR2A binding to chloroplasts. In fact, sHsp17.8 enhances AKR2A binding to chloroplasts through its interaction with both AKR2A and chloroplasts. AKR2A may also have an interaction with protein factor(s) at the COM at the insertion step of its client proteins. Import channel Toc75 has been shown to assist insertion of proteins to the COM in vitro (Bae et al., 2008; Lee et al., 2014; Tu et al., 2004). The physiological significance of the PG and MGDG binding to the ARD is demonstrated in planta using the mgd1 and pgp1-1 mutants that have a defect in biosynthesis of MGDG and PG, respectively, and consequently exhibit the yellow leaf phenotype with greatly reduced levels of COM proteins. Moreover, we provide multiple lines of evidence that these lipids, PG in particular, are present in the outer leaflet of the COM and directly involved in binding to AKR2A. Based on these findings, we propose that MGDG and PG function as the receptor for AKR2A. Lipids are known to function as a site-specific marker in recruitment of proteins to a particular membrane in diverse biological processes (Di Paolo and De Camilli, 2006; McLaughlin et al., 2002). Although the unique lipid composition of the COM was reported to be essential for the correct topology of AtOEP7 (Schleiff et al., 2001), it has not been shown that lipids can directly function as the receptor of the targeting factor. In SRP-mediated protein targeting to the ER, the protein factor SR serves as the receptor for SRP (Keenan et al., 2001). Of the two lipids, MGDG is a major lipid component of the COM and also unique to chloroplast membranes (Block et al., 1983). Thus, it can serve as a marker specific for chloroplasts among various organelles. Interestingly, however, ARD did not bind to MGDG alone. PG binding is prerequisite for the MGDG binding of the ARD, suggesting that PG binding primes MGDG binding. PG is another abundant lipid in the COM (Block et al., 1983) but it has not been implicated in protein targeting to chloroplasts. The structural, mutational and computational studies on the ARD identify two neighboring binding sites for PG (L2 site) and MGDG (L1 site). PG can bind both L1 and L2 sites in the absence of MGDG. However, the PG binding alone may not provide enough specificity for targeting of chloroplast proteins. Indeed, PG is also found in mitochondria (Douce, 1985). Thus, coincident binding of PG and MGDG to their respective sites is an elaborate mechanism to ensure tighter and more specific binding. Indeed, coincident recognition of two separate receptor molecules has recently emerged as an important mechanism for targeting proteins to specific organelles and cell membranes (Di Paolo and De Camilli, 2006). However, a previous study showed that Toc34 is directly inserted into protein-free liposomes in vitro (Qbadou et al., 2003), raising the possibility that COM targeting and insertion processes are controlled by different mechanisms.
This type of interaction between the ARD and its receptor consisting of two lipids at the target organelle may be an ideal mechanism for recruiting AKR2A to the COM in AKR2A-mediated protein targeting to chloroplasts. The cytosolic targeting factor should be released rapidly from the target membrane after delivering cargo proteins to the target membrane. In the case of SRP in the ER targeting, its interaction with SR is regulated by the GTPase activity of these proteins (Keenan et al., 2001). When two different types of lipids are simultaneously and synergistically involved in protein interaction, overall binding can be easily reversed by disrupting one of two interactions, PG or MGDG binding. Thus, the coincidental and synergistic interaction of PG and MGDG with the ARD could be ideally suited for both high specificity and tight regulation. It is not known at present how the interaction between the two lipids and the ARD can be regulated. One can speculate that a conformational change of ARD caused by a post-translational modification or a change in local concentration in PG and MGDG may serve as a regulatory mechanism.
Evolution of the AKR2A-mediated protein targeting mechanism during organellogenesis of the chloroplast
Establishment of the protein targeting mechanisms to chloroplasts should have been one of the most pivotal processes in conversion of endosymbiotic cyanobacterium to the chloroplast during evolution (Cavalier-Smith and Lee, 1985; Kim and Hwang, 2013). The most compelling evidence that the ER originated from the plasma membrane was provided by the SRP-mediated protein targeting to the ER (Warren and Wickner, 1996). Likewise, the mechanism of protein targeting to the chloroplasts may provide a clue to the question of how endosymbiotic cyanobacterium were converted chloroplasts during evolution. We provide convincing evidence that AKR2A evolved from an ARD of the host cell. To acquire the ability to function as the cytosolic targeting factor, the ARD appears to have evolved in two directions. One is related to the cargo binding activity and regulation of protein stability. The N-terminal region of AKR2A appears to be generated by a sequential addition of three conserved domains. The C1 and C2 domains, which are involved in the cargo binding, appear to be added through at least two steps before the divergence of plants from algae. In contrast, the N-terminal PEST sequence, which is involved in regulation of protein stability, seems to be added after divergence of plants from algae. The PEST sequence is a well-known motif involved in regulation of protein activity (Rechsteiner and Rogers, 1996). It is possible that the PEST sequence was added to multi-cellular plant proteins, but not to single-celled algal proteins, for cell type-specific regulation of AKR2A.
Another evolutionary direction of the ARD evolution is related to the recognition of the target organelle, chloroplasts. Our results suggest that the ARD evolved from a protein-protein interaction module or structural module into a lipid binding domain. Orthologs of AKR2A exist in diverse plant species. Furthermore, the AKR2A-type ARD containing proteins, AKR2R and AKR2L, exist in various algae. Among these proteins, key lipid binding residues are evolutionary well conserved. Interestingly, these residues are not conserved in ankyrin repeats of cyanobacterial and animal proteins. Thus, it appears that the membrane binding activity of the ARD of AKR2A was acquired in the host cell where the endosymbiotic cyanobacterium was undergoing organellogenesis to achieve specific chloroplast targeting, hence successful endosymbiosis of chloroplasts during evolution.
In addition, this study provides another important clue for the evolution of the targeting mechanism. Of the two lipids, PG and MGDG, which function as the receptor of AKR2A, MGDG is thought to be a remnant of the endosymbiotic cyanobacterium. In addition, the high amounts of PG in chloroplast membranes might have resulted from the lipid composition of the progenitor, the endosymbiotic cyanobacterium. Thus, two abundant lipids in the outer membrane of the cyanobacterium were selected as a specific target of the endosymbiotic cyanobacterium which was undergoing organellogenesis into the chloroplast, and the ARD in the host cell evolved into a lipid binding domain so as to recognize these two lipids. Based on these findings, we propose that the core components of the mechanism of chloroplast protein targeting derived from both the host cell and endosymbiotic cyanobacterium. Collectively, our study provides an important new clue to how the protein targeting mechanism of chloroplast proteins was established during organellogenesis of the chloroplast.
EXPERIMENTAL PROCEDURES
Growth of plants
Arabidopsis plants were grown on soil or MS plates supplemented with 1% sucrose in a growth chamber at 20 – 22°C under a 16 h/8 h light/dark cycle. Leaf or whole tissues were harvested from 10-day- to 3-week-old plants and used immediately for protoplast isolation, chlorophyll extraction or total RNA extraction.
Construction of plasmid DNAs
The constructs of His-tagged recombinant proteins of various ARD alanine substitution mutants were generated by a PCR approach. The details of the plasmid construction are present in Supplemental Experimental Procedures. Construction of His:AKR2A, His:ARD(211–342), His:ARD(211–309), His:ARD(211–276), His:ARD(244–342), His:ARD(277–342), His:sHsp17.8, AKR2A:HA, GFP:AtOEP7, AtOEP7:GFP, and AtToc64:GFP has been described previously (Bae et al., 2008; Kim et al., 2011; Lee et al., 2001). All PCR products were sequenced to confirm the nucleotide sequences.
Transient expression of proteins in protoplasts
The fusion constructs were transformed into protoplasts by polyethylene glycol-mediated transformation as described previously (Jin et al., 2001; Kim et al., 2001). Expression of transiently expressed proteins was examined either by Western blot analysis or in vivo imaging.
GFP signals in protoplasts were monitored at various times after transformation, and images were captured with a Zeiss LSM 510 META laser scanning confocal microscope (Zeiss) using a C-APOCHROMAT (× 40/1.2w numerical aperture water immersion) lens in a multitrack mode. Excitation/emission wavelength was 488/505 to 530 nm for GFP.
For fractionation of GFP:AtOEP7, the protoplast lysates were fractionated into soluble and pellet fractions by centrifugation at 1,500 × g at 4°C for 10 min. The soluble fractions were separated again into soluble and pellet fractions by ultracentrifugation at 100,000 × g at 4°C for 1 h. The pellet fractions were resuspended to the original volume of the lysis buffer [50 mM Tris-HCl, pH 7.5, 15 mM NaCl, 3 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT and 1 × complete protease inhibitor cocktail (Roche)]. These two fractions were analyzed by western blotting.
Lipid vesicles preparation and SPR analysis
Small unilamellar vesicles were prepared using a Liposofast (Avestin) microextruder with a 100 nm polycarbonate filter. All SPR measurements were performed at 23°C in 20 mM Tris-HCl, pH 7.4, containing 0.16 M NaCl using a lipid-coated L1 chip in the BIACORE T100 system (GE healthcare, Sweden). POPC/POPG/MGDG (varying ratios) vesicles and POPC vesicles were coated onto the active surface and the control surface, respectively. Vesicles were coated onto the corresponding sensor chip surfaces to yield the identical resonance units (RU), ensuring the equal concentration of the coated lipids. Equilibrium SPR measurements were done at the flow rate of 5 µl/min to allow sufficient time for the response in the association phase to reach near-equilibrium values (Req) (Ananthanarayanan et al., 2003). A minimum of 5 different protein concentrations were injected to collect a set of Req values that were plotted against the protein concentrations (Po). An apparent dissociation constant (Kd) was then determined by nonlinear least squares analysis of the binding isotherm using the following equation, Req = Rmax /(1 + Kd/Po) where Rmax indicates the maximal Req value (Cho et al., 2001). Since the concentration of lipids coated on the sensor chip cannot be accurately determined, Kd is defined in terms of Po as Po yielding half-maximal binding with a fixed lipid concentration. The measurement was repeated at least three times to determine average and standard deviation values. Kinetic SPR measurements were performed at the flow rate of 30 µl/min.
Protein expression and purification
E. coli BL21(DE3) cells transformed with various expression constructs were cultured to an OD600 of ~0.6. Expression of the encoded proteins was induced by adding 0.2 – 1 mM IPTG at 37°C for 3 h. His-tagged recombinant proteins were purified using an Ni+-NTA affinity column (Qiagen) according to the manufacturer’s protocol. E. coli extracts containing the recombinant proteins were incubated with Ni+-NTA agarose bead and washed several times with washing buffer [50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole, 1% Triton X-100, 1 × complete protease inhibitor cocktail (Roche)]. For crystallization of ARD proteins, His:ARD was further purified by anion exchange (MonoQ) and gel-filtration chromatography (Superdex 75), concentrated to 10 mg/ml by ultrafiltration and stored at −80°C.
Crystallization and data collection
Crystals of the ARD was grown at 22°C by the hanging drop vapor diffusion method. The crystallization buffer contained 30% PEG4K, 0.1 M Tris-HCl, pH 8.5, 0.2 M Magnesium Chloride hexahydrate, and 5% acetone. Diffraction data were collected at −170°C using crystals flash-frozen in crystallization buffer containing 30% (w/v) glycerol. Diffraction data from native crystals were collected at 1.000 Å on a Beamline 4A at the Pohang Advanced Light Source. The ARD crystals formed in space group P21 (a = 68.3 Å, b = 59.4 Å. c = 84.9 Ǻ, and β=107.3°) and contained five ARD molecules in an asymmetric unit (PDB ID: 4TUM). Diffraction data integration, scaling, and merging were performed using the HKL2000 package (Otwinowski and Minor, 1997).
Structure determination and refinement
The structure of the ARD was determined by the molecular replacement method. We determined the structure of ARD with the MolRep program using 4ANK (1N0R_PDB code) as a search model. We have identified five ARD molecules in an asymmetric unit, and have trace most of the chains. Successive rounds of model building using COOT (Emsley and Cowtan, 2004) and refinement using PHENIX program (Adams et al., 2010) dropped the R-free to 22.4%. The final model consists of residues 219–340 of AKR2A (4TUM_PDB code). The N- and C-terminal residues were not visible and not modeled.
Molecular modeling
To comprehensively evaluate all possible AKR2A-ligands complexes, we initially used the complete systematic search program PatchDock (http://bioinfo3d.cs.tau.ac.il/PatchDock) to dock a head group of PG and/or MGDG to ARD. The coordinates of MGDG and PG were taken from the PDB structures 3arc and 3mtx, respectively. Before the docking procedure, the fatty acid chains of the lipids were deleted up to the methyl groups. In all 10 of the analyzed highest scoring solutions, MGDG primarily binds to the L1 site, whereas PG mainly binds to the L2 and less frequently to the L3 site.
Docking of the lipids to the structure of ARD was performed with the package DOCK 6 (http://dock.compbio.ucsf.edu/DOCK_6/index.htm). The spheres representing the binding site were selected within 10 Å from the side-chains of the residues shown to be important for the lipid binding (H223 and E246 for MGDG, and Y261, Y290, Y294, and R296 for PG). After the docking, each aliphatic tail was extended to three carbons using the structural superimposition with the initial lipid ligands and subsequent minimization of the structure. Superimposition and minimization were performed with the UCSF Chimera package (Pettersen et al., 2004).
Statistical analysis
Data are expressed as mean values ± standard deviation (SD). No statistical method was used to predetermine sample sizes. Statistical significance was performed using the Student’s 2 tail t-test. A p-value <0.05 was considered statistically significant. In all quantitative figures * means p-value <0.05, ** p-value <0.01, *** p-value <0.001.
Supplementary Material
Highlights.
Ankyrin repeat domain recognizes coincidentally and synergistically MGDG and PG.
Structural and mutational studies identify two adjacent PG and MGDG binding sites.
AKR2A evolved from the ARD of the host cell.
MGDG and PG of endosymbiotic cyanobacteria were selected as the receptors of AKR2A.
ACKNOWLEDGEMENTS
We thank Christoph Benning (Michigan State University, USA) for providing the pgp1-1 mutant seeds and Huan Qiu (Rutgers University, USA) for assistance in bioinformatics. This work was supported in part by grants from the World Class University program (R31-2011-000-10105-0, I.H. and W.C.), the National Research Foundation (20120001015, I.H.; 2012004028 and 20100019706, Y.C.), and Global Frontier program (20110031340, I.H.) of the Ministry of Education, Science and Technology (Korea), a rising star program, POSTECH (Y.C.), Korean RDA grant (BioGreen21 PJ008177, H.S.Y) and the National Institutes of Health GM68849 (W.C.) and GM-30518 and GM-094597 (B.H.).
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta. Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthanarayanan B, Stahelin RV, Digman MA, Cho W. Activation mechanisms of conventional protein kinase C isoforms are determined by the ligand affinity and conformational flexibility of their C1 domains. J. Biol. Chem. 2003;278:46886–46894. doi: 10.1074/jbc.M307853200. [DOI] [PubMed] [Google Scholar]
- Babiychuk E, Müller F, Eubel H, Braun HP, Frentzen M, Kushnir S. Arabidopsis phosphatidylglycerophosphate synthases 1 is essential for chloroplast differentiation, but is dispensable for mitochondrial function. Plant J. 2003;33:899–909. doi: 10.1046/j.1365-313x.2003.01680.x. [DOI] [PubMed] [Google Scholar]
- Bae W, Lee YJ, Kim DH, Lee J, Kim S, Sohn EJ, Hwang I. AKR2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nature Cell Biol. 2008;10:220–227. doi: 10.1038/ncb1683. [DOI] [PubMed] [Google Scholar]
- Barrick D, Ferreiro DU, Komives EA. Folding landscapes of ankyrin repeat proteins: experiments meet theory. Curr. Opin. Struct. Biol. 2008;18:27–34. doi: 10.1016/j.sbi.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block MA, Dorne AJ, Joyard J, Douce R. Preparation and characterization of membrane fractions enriched in outer and inner envelope membranes from spinach chloroplasts. II. Biochemical characterization. J. Biol. Chem. 1983;258:13281–13286. [PubMed] [Google Scholar]
- Cavalier-Smith T, Lee JJ. Protozoa as hosts for endosymbioses and the conversion of symbionts into organelles. J Protozool. 1985;32:376–379. [Google Scholar]
- Chen X, Schnell DJ. Protein import into chloroplasts. Trends Cell Biol. 1999;9:222–227. doi: 10.1016/s0962-8924(99)01554-8. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sheng R, Kallberg M, Silkov A, Tun MP, Bhardwaj N, Kurilova S, Hall RA, Honig B, Lu H, Cho W. Genome-wide functional annotation of dual specificity protein- and lipid-binding modules that regulate protein interactions. Molecular Cell. 2012;46:226–237. doi: 10.1016/j.molcel.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W, Bittova L, Stahelin RV. Membrane binding assays for peripheral proteins. Anal. Biochem. 2001;296:153–161. doi: 10.1006/abio.2001.5225. [DOI] [PubMed] [Google Scholar]
- Cho W, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys Biomol. Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- Dhanoa PK, Richardson LG, Smith MD, Gidda SK, Henderson MP, Andrews DW, Mullen RT. Distinct pathways mediate the sorting of tail-anchored proteins to the plastid outer envelope. PLoS One. 2010;5:e10098. doi: 10.1371/journal.pone.0010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Dorne AJ, Joyard J, Block MA, Douce R. Localization of phosphatidylcholine in outer envelope membrane of spinach chloroplasts. J Cell Biol. 1985;100:1690–1697. doi: 10.1083/jcb.100.5.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce R. Mitochondria in higher plants: structure, function and biogenesis. New York: Academic Press Inc.; 1985. p. 322. [Google Scholar]
- Dyall SD, Brown MT, Johnson PJ. Ancient invasions: from endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Finet C, Timme RE, Delwiche CF, Marletaz F. Multigene phylogeny of the green lineage reveals the origin and diversification of land plants. Current Biology. 2010;20:2217–2222. doi: 10.1016/j.cub.2010.11.035. [DOI] [PubMed] [Google Scholar]
- Flores-Pérez U, Jarvis P. Molecular chaperone involvement in chloroplast protein import. Biochim. Biophys. Acta. 2013;1833:332–340. doi: 10.1016/j.bbamcr.2012.03.019. [DOI] [PubMed] [Google Scholar]
- Girzalsky W, Saffian D, Erdmann R. Peroxisomal protein translocation. Biochim. Biophys. Acta. 2010;1803:724–731. doi: 10.1016/j.bbamcr.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Hachmann AB, Angert ER, Helmann JD. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob Agents Chemother. 2009;53:1598–609. doi: 10.1128/AAC.01329-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachmann AB, Sevim E, Gaballa A, Popham DL, Antelmann H, Helmann JD. Reduction in membrane phosphatidylglycerol content leads to daptomycin resistance in Bacillus subtilis. Antimicrob Agents Chemother. 2011;55:4326–4337. doi: 10.1128/AAC.01819-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann NR, Theg SM. Chloroplast outer membrane protein targeting and insertion. Trends Plant Sci. 2005;10:450–457. doi: 10.1016/j.tplants.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Jackson DT, Froehlich JE, Keegstra K. The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J. Biol. Chem. 1998;273:16583–16588. doi: 10.1074/jbc.273.26.16583. [DOI] [PubMed] [Google Scholar]
- Jarvis P, Dörmann P, Peto CA, Lutes J, Benning C, Chory J. Galactolipid deficiency and abnormal chloroplast development in the Arabidopsis MGD synthase 1 mutant. Proc. Natl. Acad. Sci. USA. 2000;97:8175–8179. doi: 10.1073/pnas.100132197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen B, Smith C, Gibson DF, Tait JF. Entropic and enthalpic contributions to annexin V-membrane binding: a comprehensive quantitative model. J Biol Chem. 2008;283:6126–6135. doi: 10.1074/jbc.M707637200. [DOI] [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I. A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell. 2001;13:1511–1526. doi: 10.1105/TPC.000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K, Cline K. Protein import and routing systems of chloroplasts. Plant Cell. 1999;11:557–570. doi: 10.1105/tpc.11.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu. Rev. Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- Kim DH, Eu YJ, Yoo CM, Kim YW, Pih KT, Jin JB, Kim SJ, Stenmark H, Hwang I. Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell. 2001;13:287–301. doi: 10.1105/tpc.13.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Hwang I. Direct targeting of proteins from the cytosol to organelles: the ER versus endosymbiotic organelles. Traffic. 2013;14:613–621. doi: 10.1111/tra.12043. [DOI] [PubMed] [Google Scholar]
- Kim DH, Xu ZY, Na YJ, Yoo YJ, Lee J, Sohn EJ, Hwang I. Small heat shock protein Hsp17.8 functions as an AKR2A cofactor in the targeting of chloroplast outer membrane proteins in Arabidopsis. Plant Physiol. 2011;157:132–146. doi: 10.1104/pp.111.178681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Kondo M, Fukuda H, Nishimura M, Ohta H. Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc. Natl. Acad. Sci. USA. 2007;104:17216–17221. doi: 10.1073/pnas.0704680104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kim DH, Hwang I. Specific targeting of proteins to outer envelope membranes of endosymbiotic organelles, chloroplasts, and mitochondria. Front Plant Sci. 2014;5:173. doi: 10.3389/fpls.2014.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Kim DH, Kim YW, Hwang I. Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell. 2001;13:2175–2190. doi: 10.1105/tpc.010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Sohn EJ, Lee KH, Lee DW, Hwang I. The transmembrane domain of AtToc64 and its C-terminal lysine-rich flanking region are targeting signals to the chloroplast outer envelope membrane [correction] Mol. Cells. 2004;17:281–291. [PubMed] [Google Scholar]
- Li J, Mahajan A, Tsai MD. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006;45:15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D. Evolutionary analysis of Arabidopsis cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S, Wang J, Gambhir A, Murray D. PIP(2) and proteins: interactions, organization, and information flow. Annu. Rev. Biophys Biomol. Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraih JK, Harris J, Taylor SD, Palmer M. Characterization of daptomycin oligomerization with perylene excimer fluorescence: stoichiometric binding of phosphatidylglycerol triggers oligomer formation. Biochim Biophys Acta. 2012;1818:673–678. doi: 10.1016/j.bbamem.2011.10.027. [DOI] [PubMed] [Google Scholar]
- Muraih JK, Pearson A, Silverman J, Palmer M. Oligomerization of daptomycin on membranes. Biochim Biophys Acta. 2011;1808:1154–1160. doi: 10.1016/j.bbamem.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Navarro J, Chabot J, Sherrill K, Aneja R, Zahler SA, Racker E. Interaction of duramycin with artificial and natural membranes. Biochemistry. 1985;24:4645–4650. doi: 10.1021/bi00338a025. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Qbadou S, Tien R, Soll J, Schleiff E. Membrane insertion of the chloroplast outer envelope protein, Toc34: constrains for insertion and topology. J Cell Sci. 2003;116:837–846. doi: 10.1242/jcs.00291. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- Reyes-Prieto A, Weber AP, Bhattacharya D. The origin and establishment of the plastid in algae and plants. Annual Review of Genetics. 2007;41:147–168. doi: 10.1146/annurev.genet.41.110306.130134. [DOI] [PubMed] [Google Scholar]
- Schleiff E, Tien R, Salomon M, Soll J. Lipid composition of outer leaflet of chloroplast outer envelope determines topology of OEP7. Mol. Biol. Cell. 2001;12:4090–4102. doi: 10.1091/mbc.12.12.4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng R, Chen Y, Yung Gee H, Stec E, Melowic HR, Blatner NR, Tun MP, Kim Y, Källberg M, Fujiwara TK, Hye Hong J, Pyo Kim K, Lu H, Kusumi A, Goo Lee M, Cho W. Cholesterol modulates cell signaling and protein networking by specifically interacting with PDZ domaincontaining scaffold proteins. Nat Commun. 2012;3:1249. doi: 10.1038/ncomms2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima M, Ohta H, Iwamatsu A, Masuda T, Shioi Y, Takamiya K. Cloning of the gene for monogalactosyldiacylglycerol synthase and its evolutionary origin. Proc. Natl. Acad. Sci. USA. 1997;94:333–337. doi: 10.1073/pnas.94.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simocková M, Holic R, Tahotná D, Patton-Vogt J, Griac P. Yeast Pgc1p (YPL206c) controls the amount of phosphatidylglycerol via a phospholipase C-type degradation mechanism. J Biol Chem. 2008;283:17107–17115. doi: 10.1074/jbc.M800868200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumandea M, Sudipto D, Sumandea C, Cho W. Roles of aromatic residues in high interfacial activity of Naja naja atra phospholipase A2. Biochemistry. 1999;38:16290–16297. doi: 10.1021/bi9921384. [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- Tu SL, Chen LJ, Smith MD, Su YS, Schnell DJ, Li HM. Import pathways of chloroplast interior proteins and the outer-membrane protein OEP14 converge at Toc75. Plant Cell. 2004;16:2078–2088. doi: 10.1105/tpc.104.023952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G, Wickner W. Organelle inheritance. Cell. 1996;84:395–400. doi: 10.1016/s0092-8674(00)81284-2. [DOI] [PubMed] [Google Scholar]
- Xu C, Härtel H, Wada H, Hagio M, Yu B, Eakin C, Benning C. The pgp1 mutant locus of Arabidopsis encodes a phosphatidylglycerolphosphate synthase with impaired activity. Plant Physiol. 2002;129:594–604. doi: 10.1104/pp.002725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Lee PJ, Kurilova S, Cho W. In situ quantitative imaging of cellular lipids using molecular sensors. Nat. Chem. 2011;3:868–874. doi: 10.1038/nchem.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.