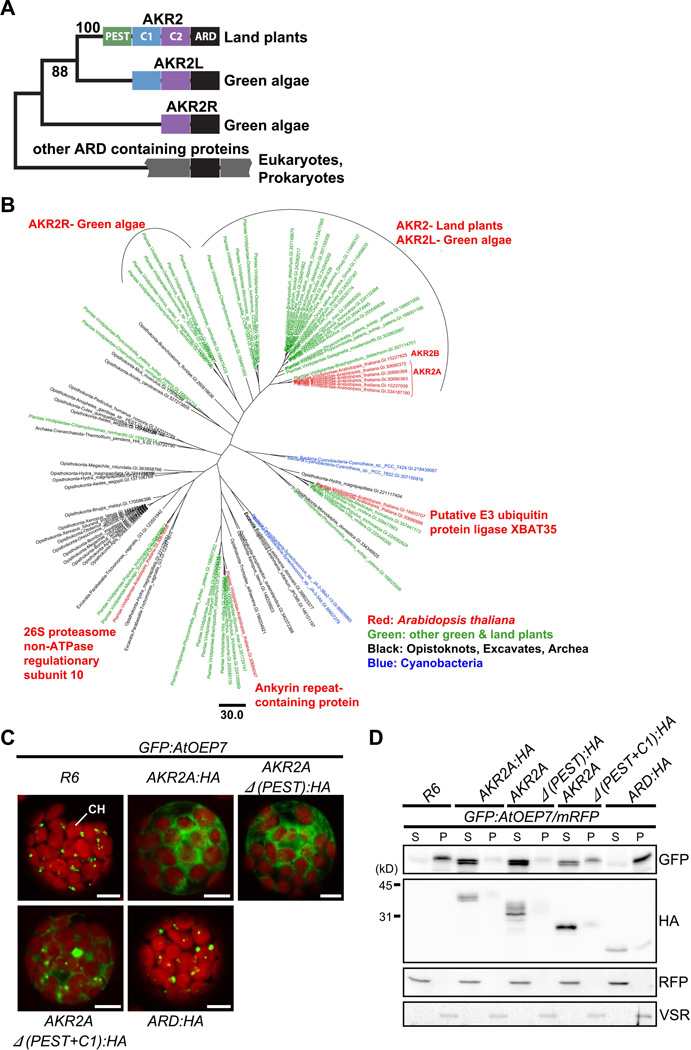
Figure 7. The Phylogenetic tree of ARDs and the evolution of AKR2A.
(A) The domain structure of ARD-containing proteins. The PEST (green), C1 (blue), C2 (purple) and ARD domains are highlighted in different colors. In cyanobacterial ARD-containing proteins, only the ARD domain (black) is shown.
(B) Maximum likelihood phylogenetic tree of ARDs. The tree is built on an alignment of 115 amino acid residues of the ARDs of 93 sequences.
(C and D) The effect of C1 and C2 domains on AKR2A binding to GFP:AtOEP7. (C) GFP:AtOEP7 was introduced into protoplasts together with HA-tagged full-length or various deletion mutants of AKR2A or empty vector R6 and the localization pattern of GFP:AtOEP7 was examined. CH, chloroplasts. Bar, 10 µm.
(D) Fractionation of GFP:AtOEP7. Protoplasts were transformed with GFP:AtOEP7 and AKR2A as in (C). mRFP was included in all transformation as a control for transformation efficiency and fractionation. Protoplast lysates were separated into soluble and pellet fractions and analyzed by Western blotting using anti-GFP, anti-HA, anti-RFP and anti-VSR antibodies. VSR was used as a control for membrane proteins. S, soluble fraction; P, pellet fraction. See also Figure S5.