Abstract
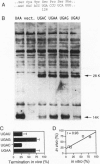
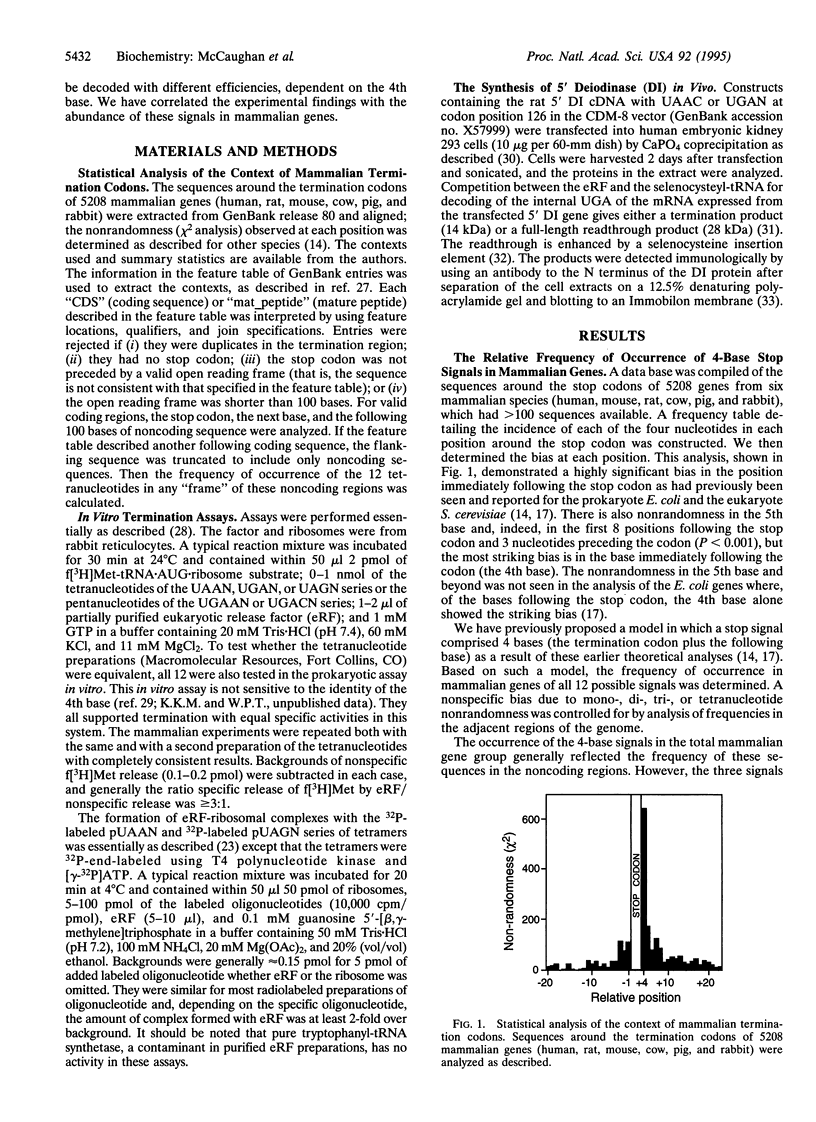
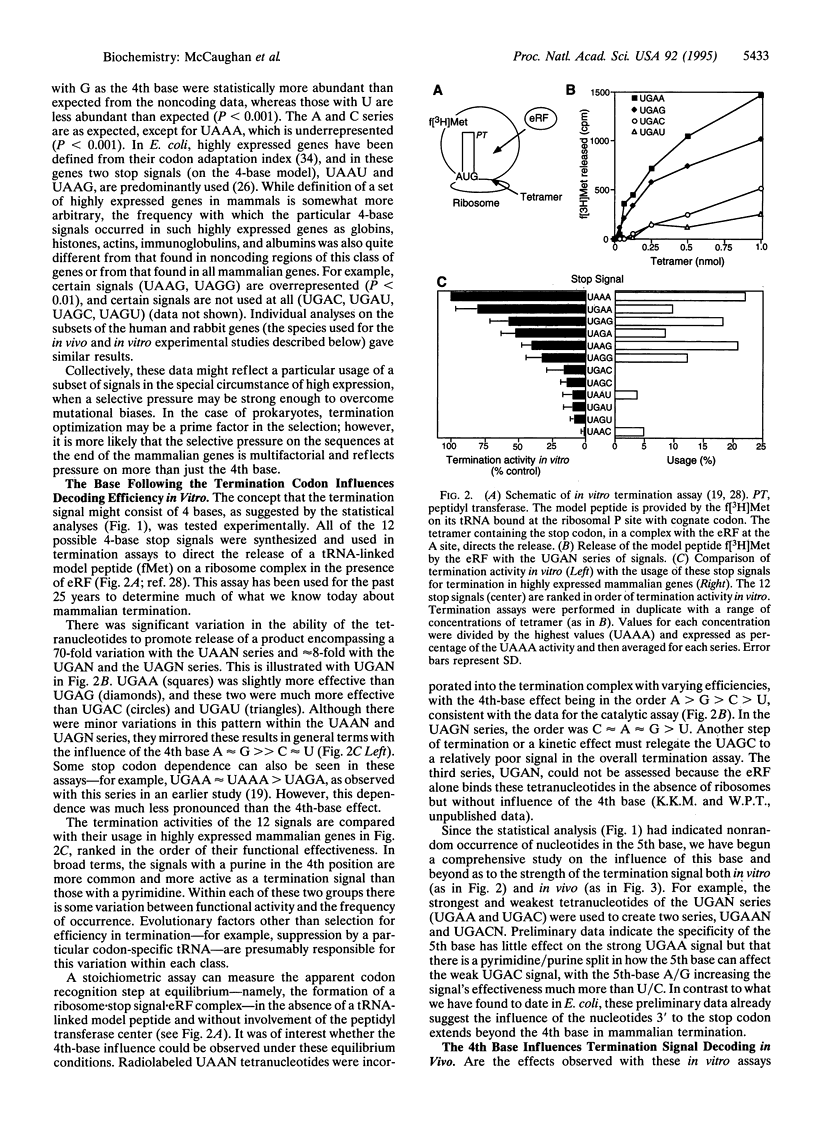
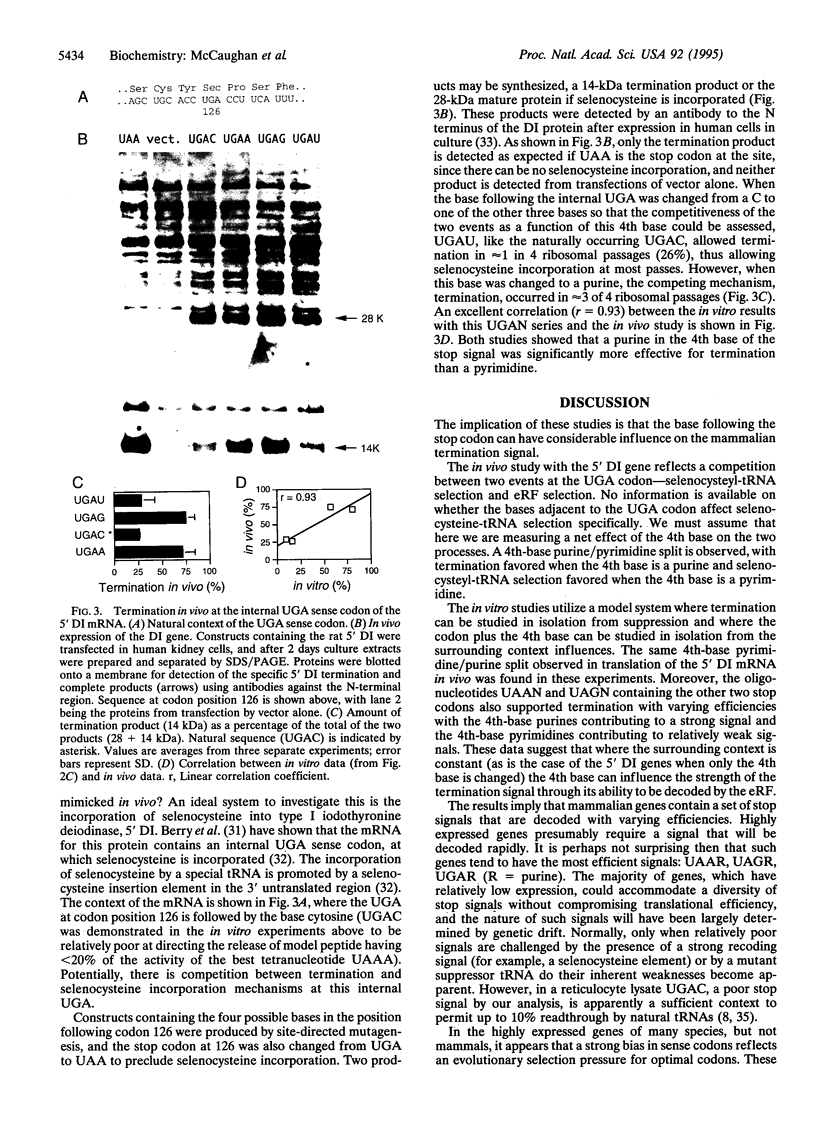
The base following stop codons in mammalian genes is strongly biased, suggesting that it might be important for the termination event. This proposal has been tested experimentally both in vivo by using the human type I iodothyronine deiodinase mRNA and the recoding event at the internal UGA codon and in vitro by measuring the ability of each of the 12 possible 4-base stop signals to direct the eukaryotic polypeptide release factor to release a model peptide, formylmethionine, from the ribosome. The internal UGA in the deiodinase mRNA is used as a codon for incorporation of selenocysteine into the protein. Changing the base following this UGA codon affected the ratio of termination to selenocysteine incorporation in vivo at this codon: 1:3 (C or U) and 3:1 (A or G). These UGAN sequences have the same order of efficiency of termination as was found with the in vitro termination assay (4th base: A approximately G >> C approximately U). The efficiency of in vitro termination varied in the same manner over a 70-fold range for the UAAN series and over an 8-fold range for the UGAN and UAGN series. There is a correlation between the strength of the signals and how frequently they occur at natural termination sites. Together these data suggest that the base following the stop codon influences translational termination efficiency as part of a larger termination signal in the expression of mammalian genes.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beaudet A. L., Caskey C. T. Mammalian peptide chain termination. II. Codon specificity and GTPase activity of release factor. Proc Natl Acad Sci U S A. 1971 Mar;68(3):619–624. doi: 10.1073/pnas.68.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M. J., Banu L., Chen Y. Y., Mandel S. J., Kieffer J. D., Harney J. W., Larsen P. R. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3' untranslated region. Nature. 1991 Sep 19;353(6341):273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- Berry M. J., Banu L., Harney J. W., Larsen P. R. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J. 1993 Aug;12(8):3315–3322. doi: 10.1002/j.1460-2075.1993.tb06001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M. J., Harney J. W., Ohama T., Hatfield D. L. Selenocysteine insertion or termination: factors affecting UGA codon fate and complementary anticodon:codon mutations. Nucleic Acids Res. 1994 Sep 11;22(18):3753–3759. doi: 10.1093/nar/22.18.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L. Context effects: translation of UAG codon by suppressor tRNA is affected by the sequence following UAG in the message. J Mol Biol. 1983 Feb 15;164(1):73–87. doi: 10.1016/0022-2836(83)90088-8. [DOI] [PubMed] [Google Scholar]
- Brent G. A., Harney J. W., Chen Y., Warne R. L., Moore D. D., Larsen P. R. Mutations of the rat growth hormone promoter which increase and decrease response to thyroid hormone define a consensus thyroid hormone response element. Mol Endocrinol. 1989 Dec;3(12):1996–2004. doi: 10.1210/mend-3-12-1996. [DOI] [PubMed] [Google Scholar]
- Brown C. M., Dalphin M. E., Stockwell P. A., Tate W. P. The translational termination signal database. Nucleic Acids Res. 1993 Jul 1;21(13):3119–3123. doi: 10.1093/nar/21.13.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. M., Stockwell P. A., Trotman C. N., Tate W. P. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 1990 Nov 11;18(21):6339–6345. doi: 10.1093/nar/18.21.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. M., Stockwell P. A., Trotman C. N., Tate W. P. The signal for the termination of protein synthesis in procaryotes. Nucleic Acids Res. 1990 Apr 25;18(8):2079–2086. doi: 10.1093/nar/18.8.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham R. H. Codon context. Experientia. 1990 Dec 1;46(11-12):1126–1133. doi: 10.1007/BF01936922. [DOI] [PubMed] [Google Scholar]
- Buckingham R. H., Sörensen P., Pagel F. T., Hijazi K. A., Mims B. H., Brechemier-Baey D., Murgola E. J. Third position base changes in codons 5' and 3' adjacent UGA codons affect UGA suppression in vivo. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):259–262. doi: 10.1016/0167-4781(90)90177-4. [DOI] [PubMed] [Google Scholar]
- Cavener D. R., Ray S. C. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991 Jun 25;19(12):3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigen W. J., Cook R. G., Tate W. P., Caskey C. T. Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donly B. C., Edgar C. D., Adamski F. M., Tate W. P. Frameshift autoregulation in the gene for Escherichia coli release factor 2: partly functional mutants result in frameshift enhancement. Nucleic Acids Res. 1990 Nov 25;18(22):6517–6522. doi: 10.1093/nar/18.22.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P. J. Alternative readings of the genetic code. Cell. 1993 Aug 27;74(4):591–596. doi: 10.1016/0092-8674(93)90507-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon K., McClendon V., Bonetti B., Bedwell D. M. Premature translation termination mutations are efficiently suppressed in a highly conserved region of yeast Ste6p, a member of the ATP-binding cassette (ABC) transporter family. J Biol Chem. 1994 Jul 8;269(27):17802–17808. [PubMed] [Google Scholar]
- Ganoza M. C., Buckingham K., Hader P., Neilson T. Effect of base sequence on in vitro protein-chain termination. J Biol Chem. 1984 Nov 25;259(22):14101–14104. [PubMed] [Google Scholar]
- Gesteland R. F., Weiss R. B., Atkins J. F. Recoding: reprogrammed genetic decoding. Science. 1992 Sep 18;257(5077):1640–1641. doi: 10.1126/science.1529352. [DOI] [PubMed] [Google Scholar]
- Kohli J., Grosjean H. Usage of the three termination codons: compilation and analysis of the known eukaryotic and prokaryotic translation termination sequences. Mol Gen Genet. 1981;182(3):430–439. doi: 10.1007/BF00293932. [DOI] [PubMed] [Google Scholar]
- Kopczynski J. B., Raff A. C., Bonner J. J. Translational readthrough at nonsense mutations in the HSF1 gene of Saccharomyces cerevisiae. Mol Gen Genet. 1992 Sep;234(3):369–378. doi: 10.1007/BF00538696. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987 Oct 26;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991 Nov;115(4):887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Rice C. M. The signal for translational readthrough of a UGA codon in Sindbis virus RNA involves a single cytidine residue immediately downstream of the termination codon. J Virol. 1993 Aug;67(8):5062–5067. doi: 10.1128/jvi.67.8.5062-5067.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. On the relationship between preferred termination codon contexts and nonsense suppression in human cells. Nucleic Acids Res. 1994 Jan 11;22(1):15–19. doi: 10.1093/nar/22.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R., Phillips-Jones M. K., Watson F. J., Hill L. S. Codon context effects on nonsense suppression in human cells. Biochem Soc Trans. 1993 Nov;21(4):846–851. doi: 10.1042/bst0210846. [DOI] [PubMed] [Google Scholar]
- Pedersen W. T., Curran J. F. Effects of the nucleotide 3' to an amber codon on ribosomal selection rates of suppressor tRNA and release factor-1. J Mol Biol. 1991 May 20;219(2):231–241. doi: 10.1016/0022-2836(91)90564-m. [DOI] [PubMed] [Google Scholar]
- Poole E. S., Brown C. M., Tate W. P. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 1995 Jan 3;14(1):151–158. doi: 10.1002/j.1460-2075.1995.tb06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Stenico M., Peden J. F., Lloyd A. T. Codon usage: mutational bias, translational selection, or both? Biochem Soc Trans. 1993 Nov;21(4):835–841. doi: 10.1042/bst0210835. [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. Quantitative analysis of the relationship between nucleotide sequence and functional activity. Nucleic Acids Res. 1986 Aug 26;14(16):6661–6679. doi: 10.1093/nar/14.16.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate W. P., Brown C. M. Translational termination: "stop" for protein synthesis or "pause" for regulation of gene expression. Biochemistry. 1992 Mar 10;31(9):2443–2450. doi: 10.1021/bi00124a001. [DOI] [PubMed] [Google Scholar]
- Tuite M. F., Stansfield I. Termination of protein synthesis. Mol Biol Rep. 1994 May;19(3):171–181. doi: 10.1007/BF00986959. [DOI] [PubMed] [Google Scholar]
- Zinoni F., Birkmann A., Stadtman T. C., Böck A. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4650–4654. doi: 10.1073/pnas.83.13.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]